Kit for identifying new coronavirus by applying mass spectrum system and using method thereof
A mass spectrometry system and kit technology, which is applied in the field of kits for the identification of new coronaviruses using mass spectrometry systems, and can solve problems such as inconvenience
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
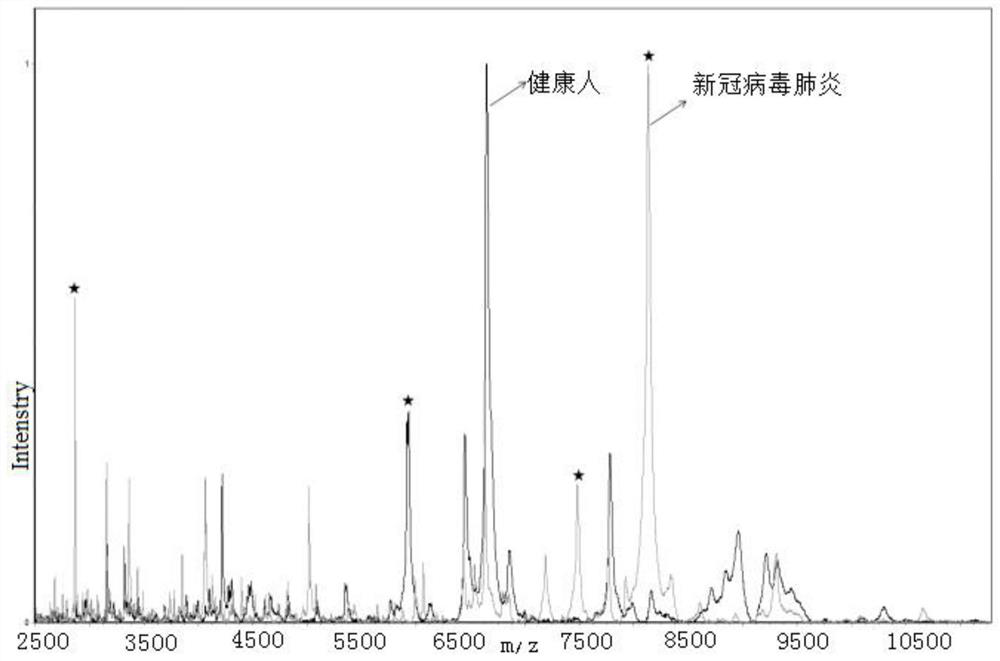
[0073] Example 1 Distinguish between novel coronavirus pneumonia and healthy people
[0074] Kit ingredients:
[0075] The preparation method of buffer U9 is: add urea and CHAPS to 0.01mol / L PBS, stir and dissolve to obtain buffer U9; the molar concentration of urea is 7mol / L, and the mass concentration of CHAPS is 8g / L.
[0076] The pH of buffer U1 is 7.5, and buffer U1 is a mixture of buffer U9 and 45mmol / L Tris-HCl.
[0077] The pH of the sodium acetate buffer is 4.0, and the molar concentration of the sodium acetate buffer is 90mmol / L.
[0078] Experimental steps:
[0079] S1 activates metalloprotein chip target plate
[0080] S2 Pretreatment of positive and negative serum samples of novel coronavirus
[0081] a) taking out the serum sample, inactivating it in a water bath after thawing; centrifuging the serum sample to separate the serum;
[0082] b) Take 20 μl of upper serum, add 30 μl of buffer U9, mix well, shake and incubate for 20 minutes to obtain intermediate ...
Embodiment 2
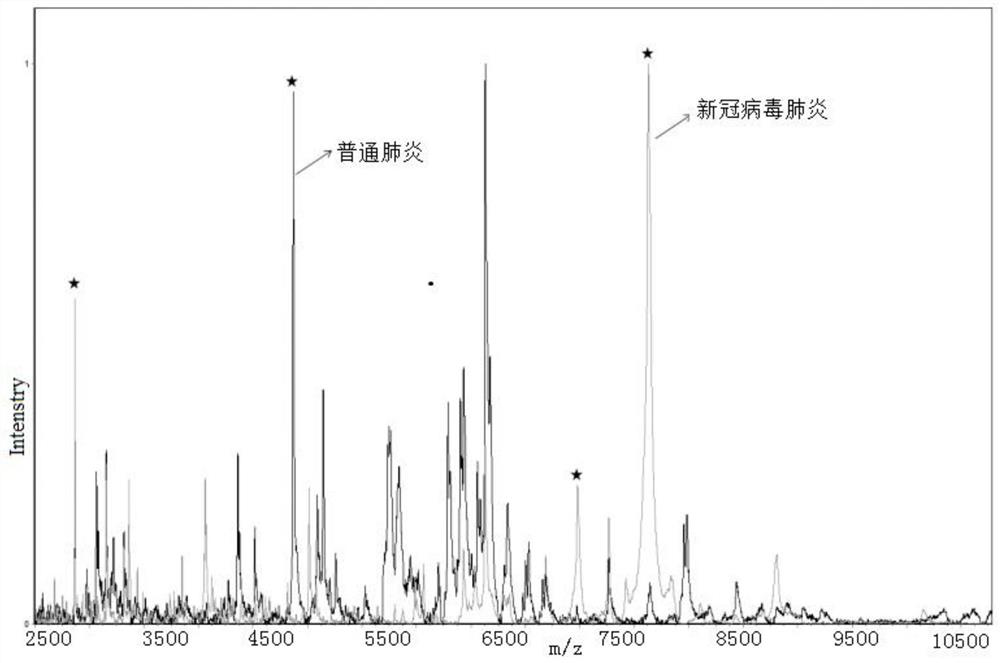
[0095] Example 2 Distinguish between novel coronavirus pneumonia and common pneumonia
[0096] Kit ingredients:
[0097] The preparation method of buffer U9 is as follows: add urea and CHAPS to 0.01mol / L PBS, stir and dissolve to obtain buffer U9; the molar concentration of urea is 8mol / L, and the mass concentration of CHAPS is 10g / L.
[0098] The pH of buffer U1 is 7.5, and buffer U1 is a mixture of buffer U9 and 50 mmol / L Tris-HCl.
[0099] The pH of the sodium acetate buffer is 4.0, and the molar concentration of the sodium acetate buffer is 105 mmol / L.
[0100] Experimental steps:
[0101] S1 activates metalloprotein chip target plate
[0102] S2 Pretreatment of positive and negative serum samples of novel coronavirus
[0103] a) taking out the serum sample, inactivating it in a water bath after thawing; centrifuging the serum sample to separate the serum;
[0104] b) Take 40 μl of upper serum, add 60 μl of buffer U9, mix well, shake and incubate for 20 minutes to obt...
Embodiment 3
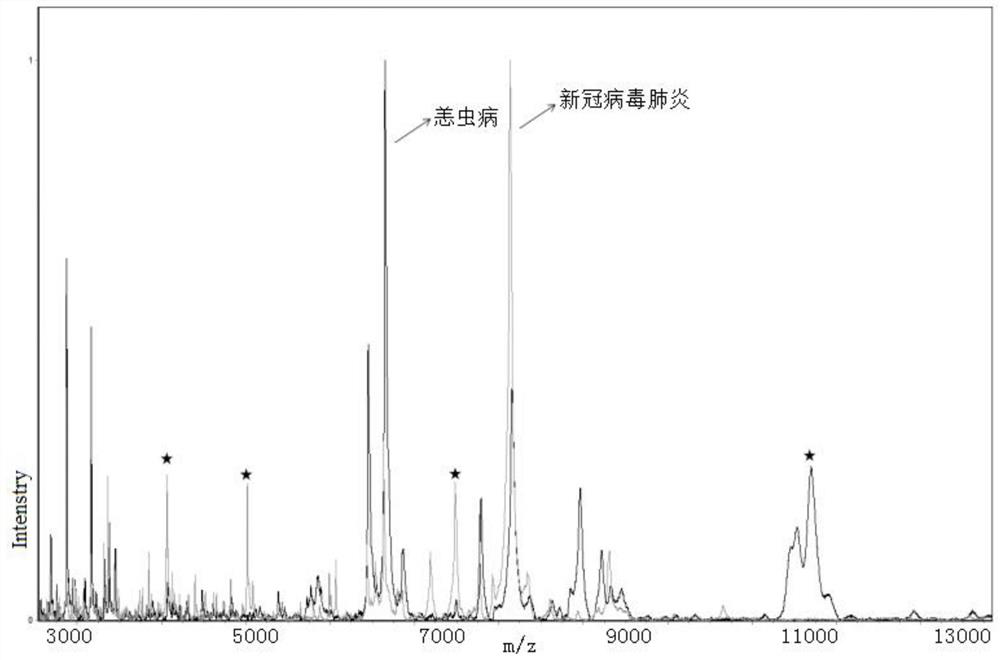
[0117] Example 3 Distinguish between novel coronavirus pneumonia and scrub typhus
[0118] Kit ingredients:
[0119] The preparation method of buffer U9 is as follows: add urea and CHAPS to 0.01mol / L PBS, stir and dissolve to obtain buffer U9; the molar concentration of urea is 10mol / L, and the mass concentration of CHAPS is 12g / L.
[0120] The pH of buffer U1 is 7.5, and buffer U1 is a mixture of buffer U9 and 60 mmol / L Tris-HCl.
[0121] The pH of the sodium acetate buffer is 4.0, and the molar concentration of the sodium acetate buffer is 120mmol / L.
[0122] Experimental steps:
[0123] S1 activates metalloprotein chip target plate
[0124] S2 Pretreatment of positive and negative serum samples of novel coronavirus
[0125] a) taking out the serum sample, inactivating it in a water bath after thawing; centrifuging the serum sample to separate the serum;
[0126] b) Take 50 μl of upper serum, add 100 μl of buffer U9, mix well, shake and incubate for 20 minutes to obtain...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com