Rapid annotation method and device for a bacterial DNA sequence
A DNA sequence and bacteria technology, applied in sequence analysis, instrumentation, genomics, etc., can solve the problems of scattered, cumbersome, and a large number of manual operations in the analysis system, and achieve the effect of simplifying and optimizing the analysis process.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 2
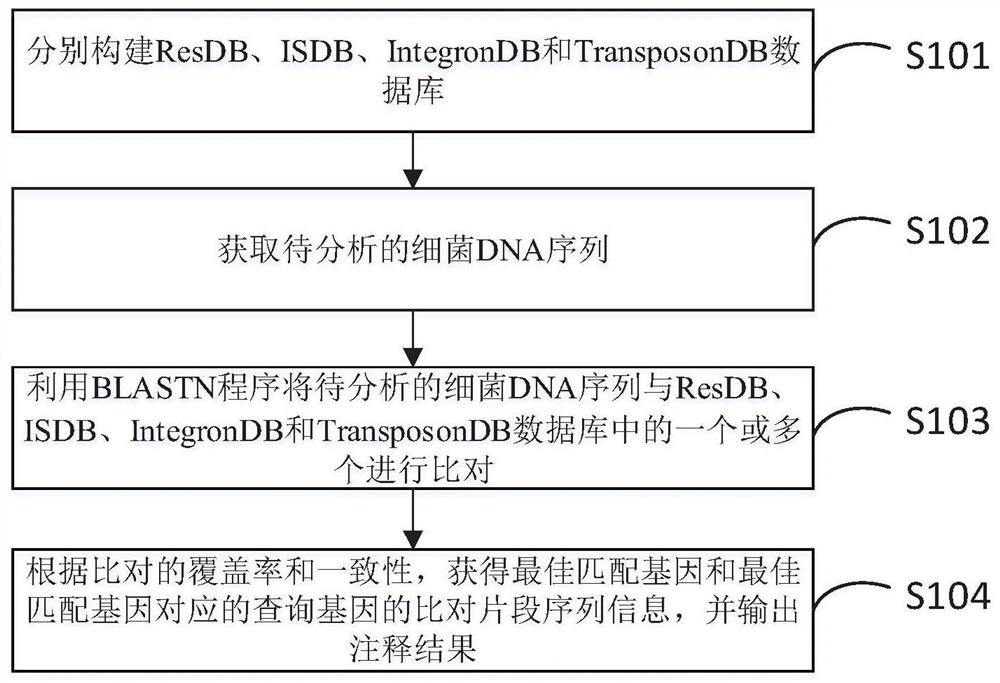
[0092] refer to Figure 4 , is a block diagram of a bacterial DNA sequence rapid annotation device provided in an embodiment of the present invention, the device can execute any of the bacterial DNA sequence rapid annotation methods provided in any embodiment of the present invention, and has corresponding functional modules for executing the method and beneficial effects. The unit includes:
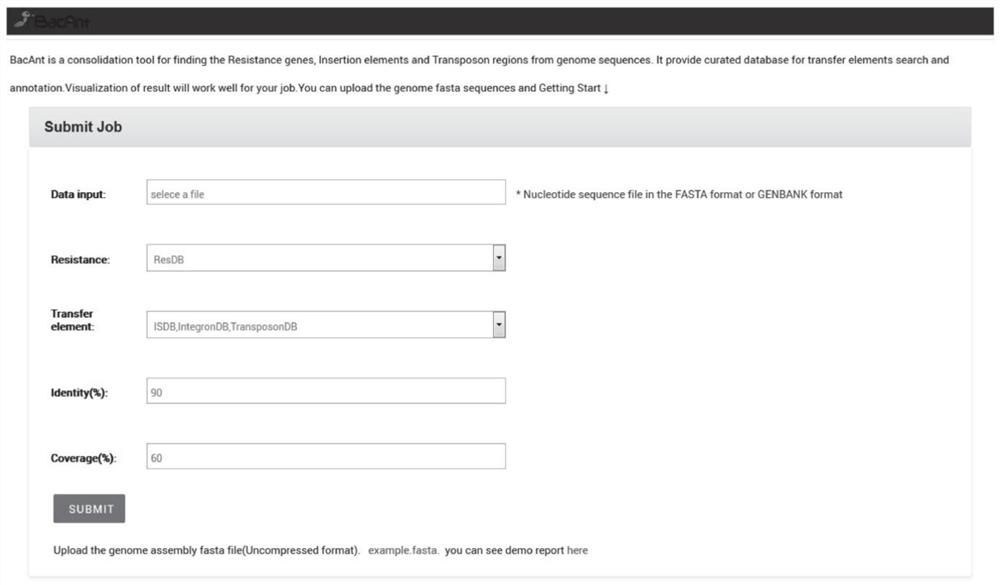
[0093] Database construction module 901, for respectively constructing ResDB, ISDB, IntegronDB and TransposonDB database;
[0094] Obtaining module 902, for obtaining the bacterial DNA sequence to be analyzed;
[0095] Comparison module 903, for utilizing the BLASTN program to compare the bacterial DNA sequence to be analyzed with one or more of ResDB, ISDB, IntegronDB and TransposonDB databases;
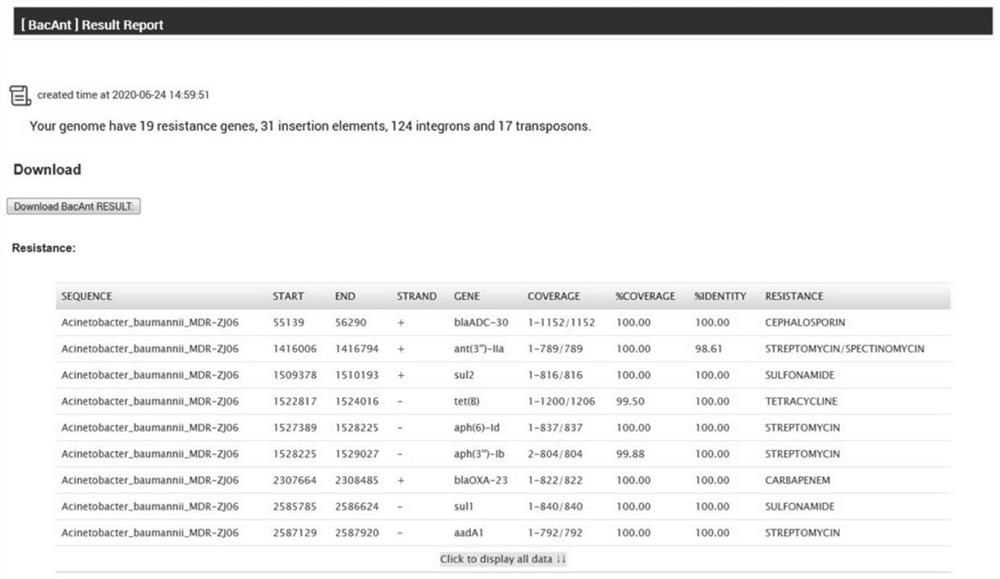
[0096] The output module 904 is configured to obtain the sequence information of the aligned fragments of the best matching gene and the query gene corresponding to the best matching gene a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com