Intron abnormal splicing repair method
A repair method and intron technology, applied in the field of genetic engineering, can solve problems such as inability to achieve lifelong cure, extremely high requirements for equipment and technology, and semi-random
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
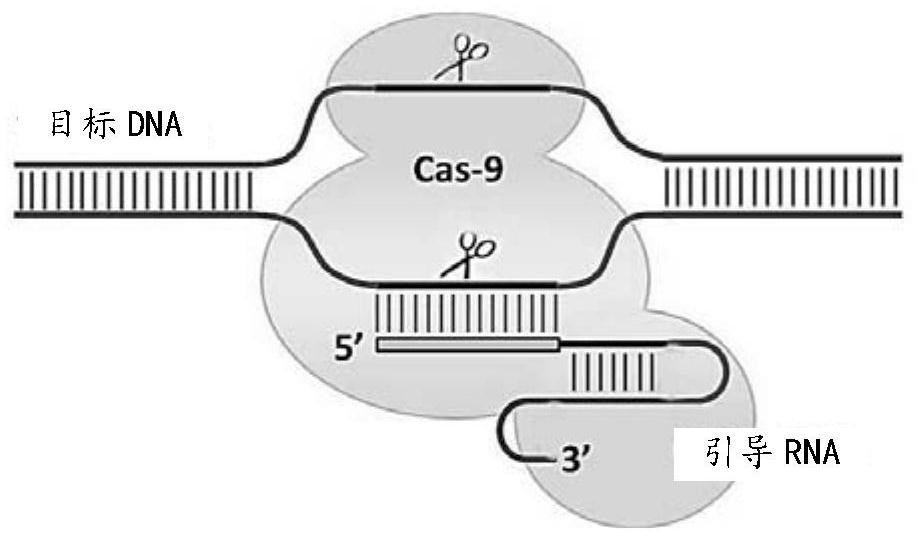
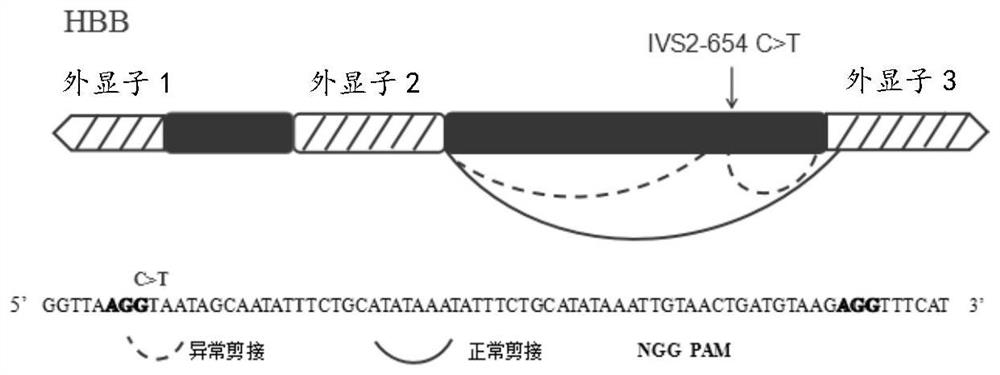
[0021]In one aspect, the present invention provides a method for repairing abnormal splicing of introns caused by the mutation of HBB (beta-globin gene) IVS2-654C>T in cells, and the IVS2-654C>T can cause additional splicing A donor site, the method comprising the step of gene editing HBB using the CRISPR-Cas9 system, the gene editing of HBB using the CRISPR-Cas9 system to inactivate the additional splicing donor site, the CRISPR-Cas9 The system includes Cas9 and at least one sgRNA targeting the target sequence.
[0022] The inactivation includes destroying the function of the above-mentioned additional splice donor site through base insertion, deletion, alteration, frameshift mutation or knockout.
[0023] Further, the targeting sequence of the sgRNA is 20 bp upstream of the IVS2-654C>T site to 70 bp downstream of the IVS2-654C>T site.
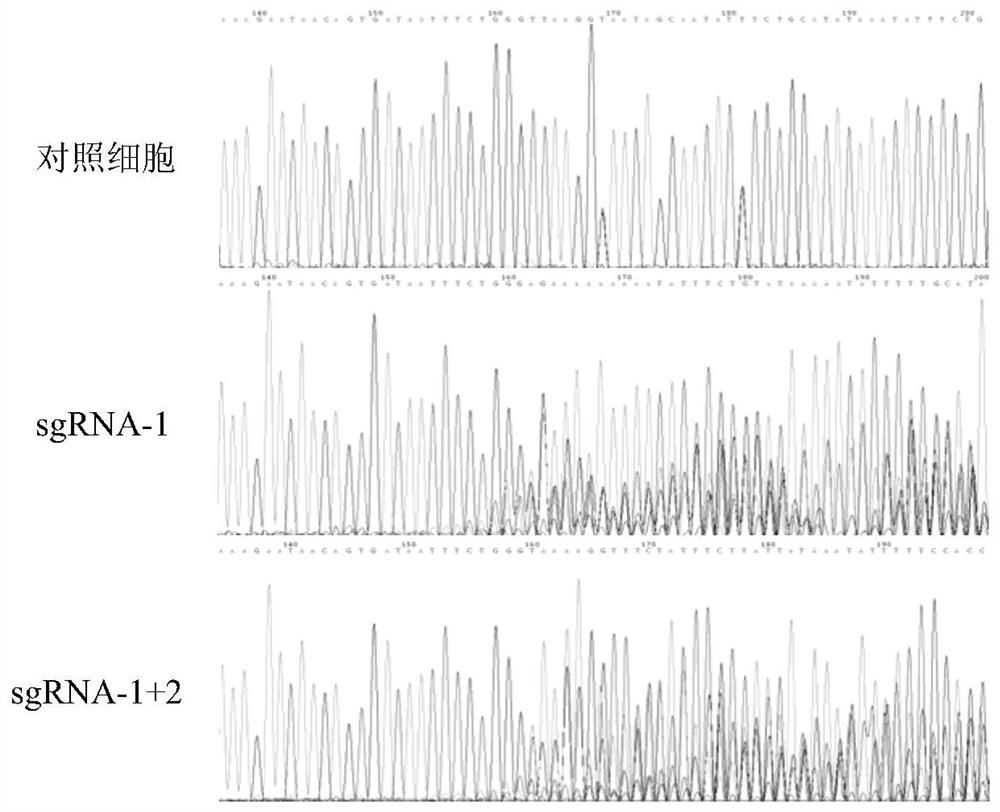
[0024] In one embodiment, the sgRNA includes sgRNA-1; in other embodiments, the sgRNA includes sgRNA-1 and sgRNA-2.
[0025] The targeting...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com