Method for improving CRISPR/Cas9 mediated biallelic mutation efficiency and application thereof
An allelic and efficient technology, applied in the fields of genetic engineering and molecular genetics and breeding, can solve problems such as the acquisition of target traits that affect the efficiency of animal gene editing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Example 1 Design of sgRNA for gene editing of MSTN and FGF5
[0039] In this example, the MSTN and FGF5 genes of sheep are taken as examples to design sgRNA for gene editing by CRISPR / Cas9.
[0040] In order to achieve high gene editing efficiency, the design principles of sgRNA are as follows: the length of the sequence complementary to the genome sequence is 20-30nt, there must be a PAM (NGG) sequence at the 3' end of the sequence, and SNP and polyT sequences should be avoided. It is best to start with the base "G".
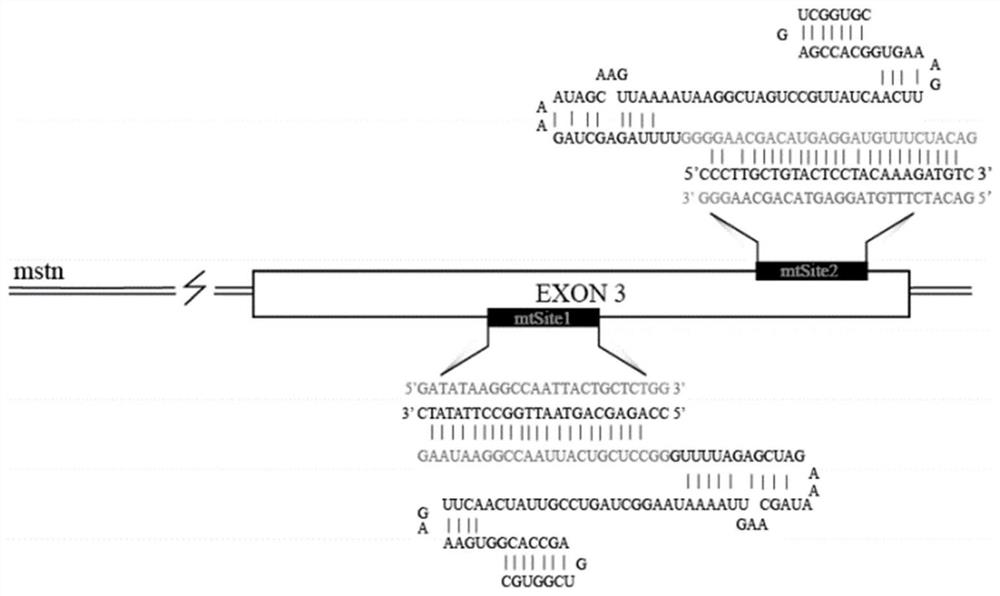
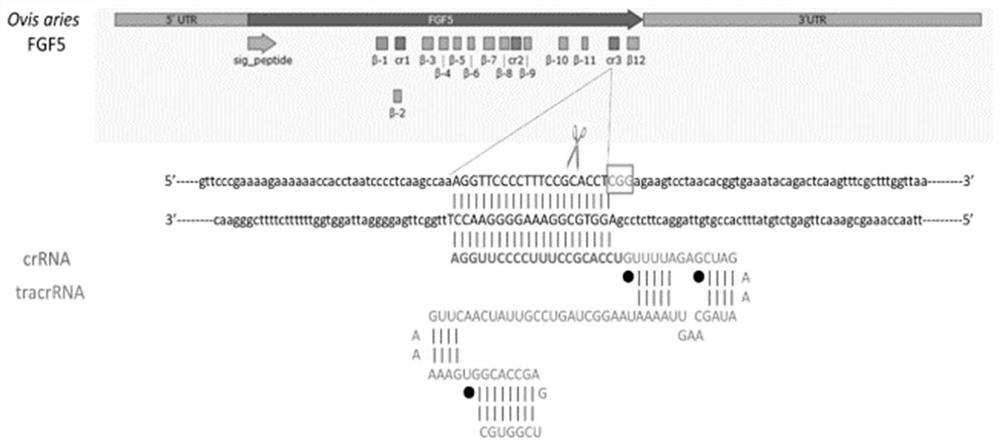
[0041] Two targeting sites were designed for the MSTN gene of sheep, both located on the third exon of the MSTN gene. The arrangement and structure of the two targets in the MSTN genome are as follows: figure 1 As shown, named as Cr1 and Cr2, figure 1The corresponding targeting sequence in the genome and the PAM segment of the site are shown. The gray sequence is the crRNA sequence corresponding to the sgRNA and the target site, followed by tracrRNA t...
Embodiment 2
[0043] Example 2 In vitro transcription of Cas9 mRNA and sgRNA
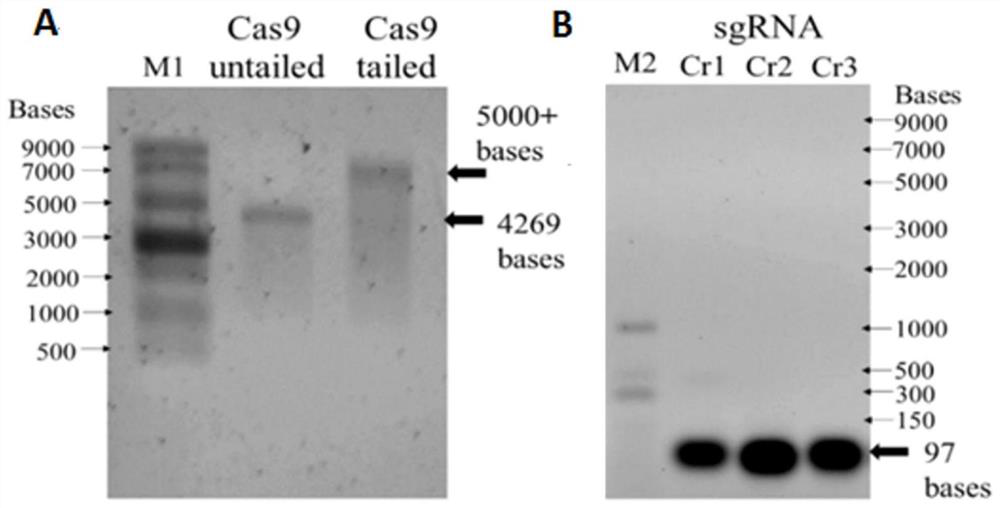
[0044] The Q5 high-fidelity DNA polymerase was used to amplify the Cas9 (Cas9 gene sequence shown in SEQ ID NO.4) and the sgRNA sequence containing the T7 promoter, respectively, and the template for in vitro transcription was obtained after purification. T7-Cas9 was transcribed with mMESSAGE mMachineT7Ultra Kit, and T7-sgRNA was transcribed with MEGAshortscript Kit. The transcribed Cas9 RNA was treated with RNA-specific loading and became a single-stranded straight line. After 2% agarose-TBE gel electrophoresis, it was shown that Cas9 was capped and transcribed at about 4200 nt. The post-transcribed Cas9 mRNA band was clear and the size was correct (such as image 3 shown in A). After the transcription of sgRNA (about 100nt) was purified and recovered, it was detected by agarose gel electrophoresis. The sgRNA band was clear and the size was correct (such as image 3 shown in B).
Embodiment 3
[0045] Example 3 Using the CRISPR / Cas9 system to edit the MSTN and FGF5 genes of sheep
[0046] In this example, the prokaryotic microinjection method was used to introduce the Cas9 mRNA synthesized by in vitro transcription and sgRNA of MSTN and FGF5 genes in Example 2 into sheep prokaryotic embryos to achieve gene knockout of MSTN and FGF5.
[0047] The Cas9 mRNA and sgRNA synthesized by in vitro transcription in Example 2 were mixed to prepare a gene editing nucleic acid solution for microinjection. According to the different injection ratios of Cas9 mRNA and sgRNA, sheep pronuclear stage embryos were divided into three groups: (1) Control group: the molar concentration ratio of Cas9 mRNA and sgRNA was 1:2, two microinjections were performed, and the first microinjection The concentrations of Cas9 mRNA and sgRNA were 5.23*10 -9 mol / l and 10.46*10 -9 mol / l, the concentrations of Cas9 mRNA and sgRNA in the second microinjection were 7.93*10 -9 mol / l and 15.86*10 -9 mol / l,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com