PCR primer for rapidly identifying weedy rice in early stage and application thereof
A weedy rice, fast technology, applied in the biological field, can solve the problems of cumbersome steps and inability to detect, and achieve the effect of simple and fast operation and accurate results.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
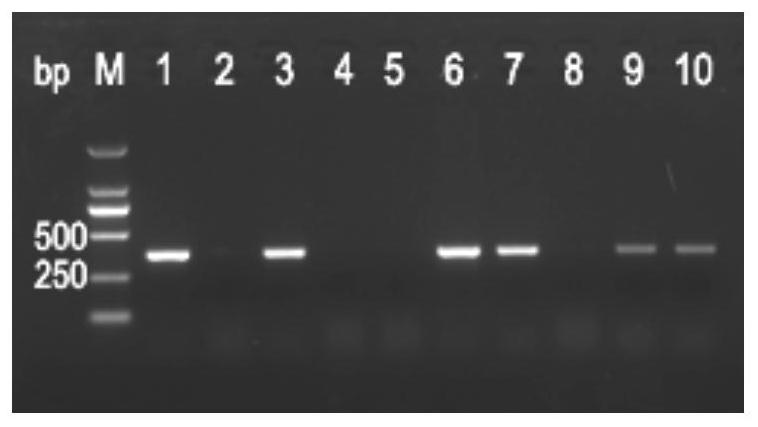
[0048]The method for rapid and early identification of weedy rice specifically includes the following steps:
[0049]1) P-Bh4 primer design
[0050]The forward primer P-Bh4-F is: 5’-TGATATGCTTCCTGCTGTGCAC-3’,
[0051]The reverse primer P-Bh4-R is: 5'-GACGGCCACGTTGAGCATCGAT-3'.
[0052]2) DNA extraction
[0053]Select 10 samples from the rice field flooded with weedy rice, collect the leaves, ground them with liquid nitrogen, and extract the genomic DNA of the samples with a plant genomic DNA extraction kit;
[0054]3) PCR amplification of sample genomic DNA
[0055]The PCR amplification reaction system is: sterile double distilled water 10.7μL, 2×PCR Mix 12.5μL, primers P-Bh4-F and P-Bh4-R (10μmol / L) each 0.4μL, template DNA (10ng / μL) ) 1 μL; amplification conditions are: 95°C pre-denaturation for 5 minutes; 95°C for 30s, 64°C for 30s, 72°C for 30s, 30 cycles; 72°C extension for 7 minutes.
[0056]4) PCR product electrophoresis detection and result judgment
[0057]After the PCR, take 7μL of the product on a ...
Embodiment 2
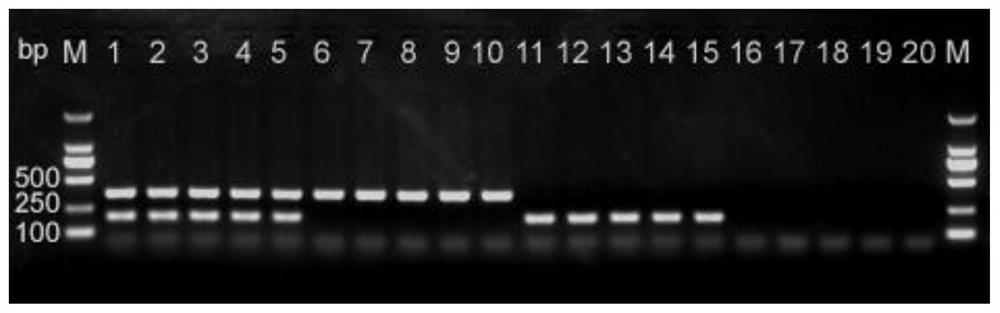
[0059]The method for quickly identifying weedy rice and cultivated rice specifically includes the following steps:
[0060]1) Primer design
[0061]a. P-Bh4 primer design:
[0062]The forward primer P-Bh4-F is: 5’-TGATATGCTTCCTGCTGTGCAC-3’,
[0063]The reverse primer P-Bh4-R is: 5'-GACGGCCACGTTGAGCATCGAT-3'.
[0064]The size of the amplified product is 356bp.
[0065]b. P-Rc primer design:
[0066]The forward primer P-Rc-F is: 5’-CATTCTCCAGATGGACTACTCC-3’,
[0067]The reverse primer P-Rc-R is: 5'-GATGGCACCGACTTTTCGCGT-3'.
[0068]The size of the amplified product is 191bp;
[0069]2) DNA extraction
[0070]120 samples of weed rice and 20 samples of cultivated rice were selected. The young leaves were collected at the seedling stage and ground with liquid nitrogen. The genomic DNA of the samples was extracted with a plant genomic DNA extraction kit;
[0071]3) Double PCR amplification of sample genomic DNA
[0072]The reaction system of double PCR amplification is: sterile double distilled water 9.5μL, 2×PCR Mix 12.5μL, p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com