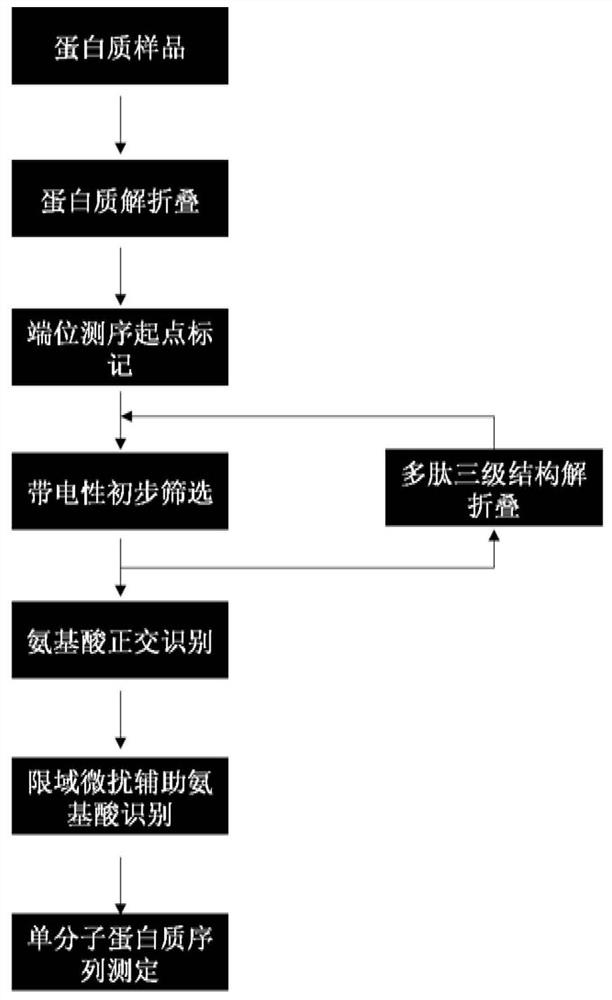
Protein/polypeptide sequencing method adopting Aerolysin nanopores
A nano-pore, protein technology, applied in the biological field, can solve the problems of inability to effectively identify 20 amino acids, difficulty in obtaining amino acid sequence information, lack of efficient organic fluorophores, etc., and achieve the effect of improving amino acid identification ability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
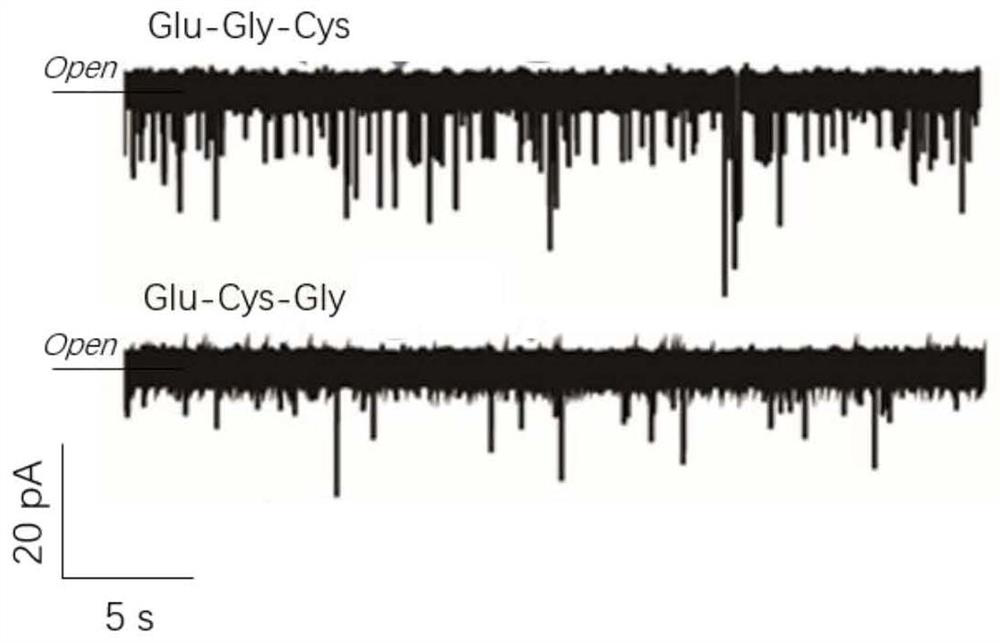
[0065] The invention discloses a method for sequencing polypeptide molecules of a cysteine-specific Aerolysin nanopore. The polypeptide uses Glu as a guide chain, and the amino acid sequences of the two polypeptide molecules are Glu-Gly-Cys and Glu-Cys-Gly respectively. Specific steps are as follows:
[0066] (1) Two electrical primary screening channels, N226Q and T232K, were designed, and the corresponding mutant Proerolysin protein was expressed and purified by site-directed mutagenesis technology for channel construction. For specific steps, refer to patent CN202010131704.8.
[0067] (2) Mix 1 mg / mL Proerolysin protein with trypsin 10:1 and incubate at room temperature for 6 hours to obtain Aerolysin monomer protein with pore-forming activity.
[0068] (3) Control the experimental temperature at 22±1°C. Add 1mL (1.0 M KCl, 10 mMTris, 1.0 mM EDTA, pH=8) buffer solution into the two detection cells respectively, and prepare the phospholipid bilayer by pulling method. For sp...
Embodiment 2
[0074] A method for detecting phosphorylated polypeptides using mutant Aerolysin nanopores, using S-K-I-G as the guide strand, the template polypeptide sequence is S-K-I-G-S-T-E-N-L, and phosphorylated modified sequences at the fifth serine and the sixth threonine respectively S-K-I-G- p S-T-E-N-L and S-K-I-G-S- p T-E-N-L. Specific steps are as follows:
[0075] (1) The wild-type electrical primary screening channel was designed, and the wild-type Proerolysin protein was expressed and purified for channel construction. For specific steps, refer to patent CN202010131704.8.
[0076] (2) Mix 1 mg / mL Proerolysin protein with trypsin 10:1 and incubate at room temperature for 6 hours to obtain Aerolysin monomer protein with pore-forming activity.
[0077] (3) Control the experimental temperature at 22±1°C. Add 1mL (1.0 M KCl, 10 mMTris, 1.0 mM EDTA, pH=8) buffer solution into the two detection cells respectively, and prepare the phospholipid bilayer by pulling method. For specif...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com