Reagent for improving replication efficiency of single-stranded nucleic acid molecule with secondary structure, nucleic acid fragment replication method and application
A single-stranded nucleic acid molecule and secondary structure technology, applied in the field of molecular biology, can solve problems such as clear mechanism and adverse effects on technical performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
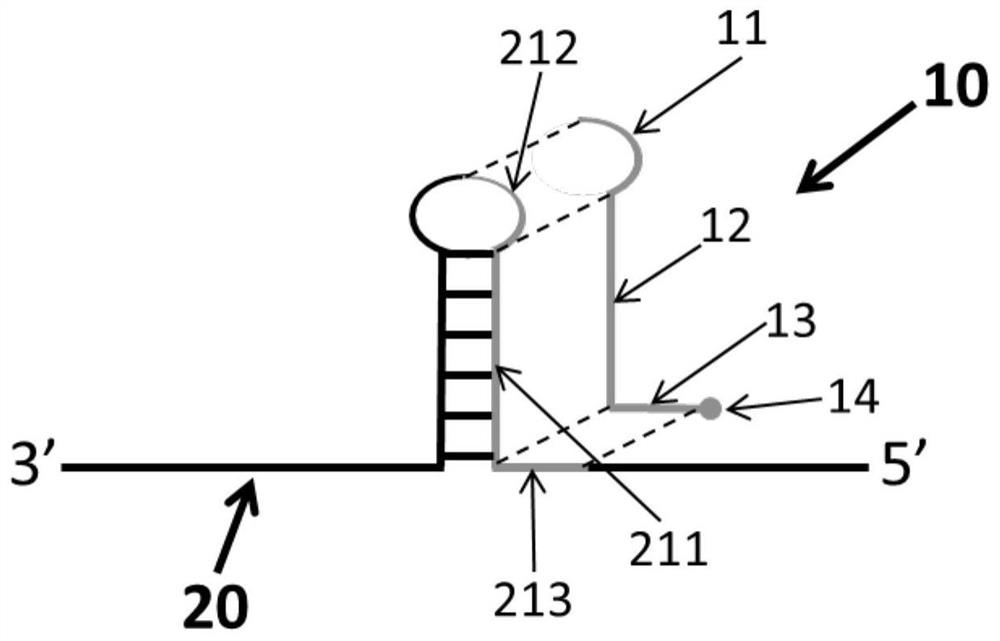
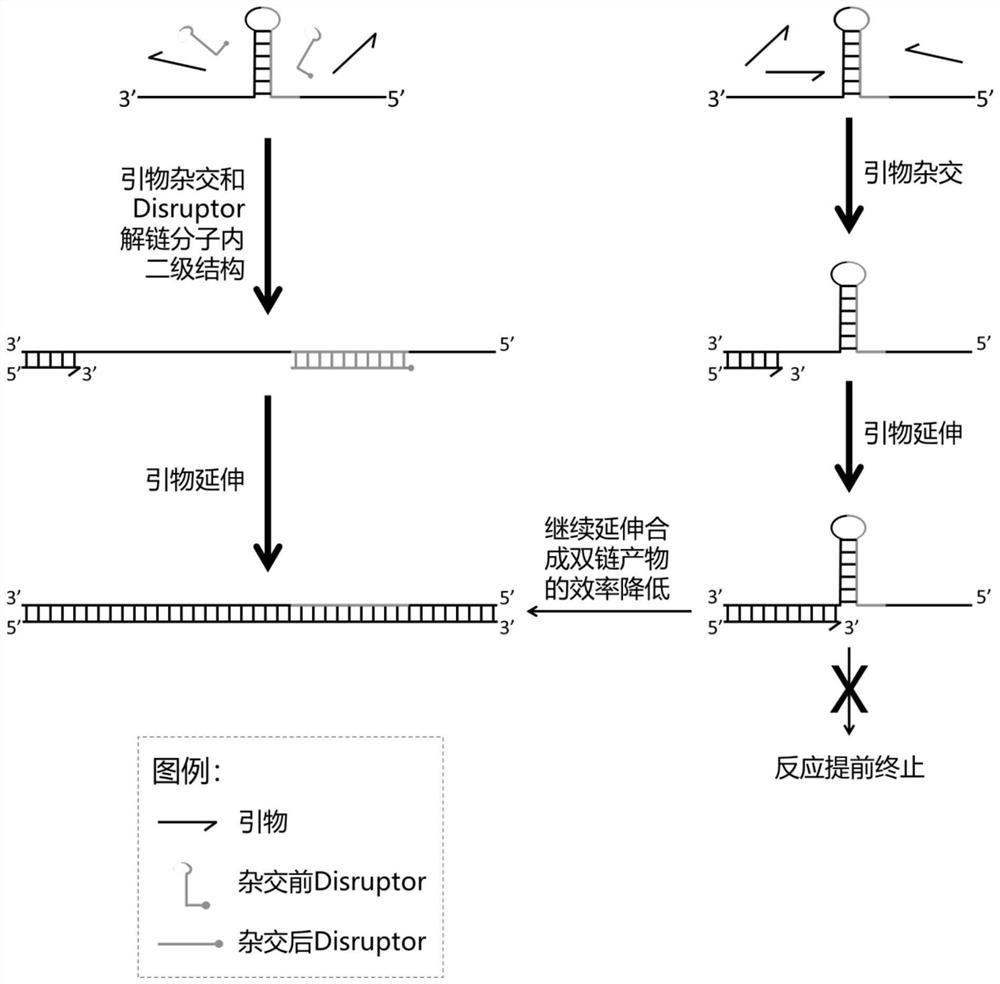
[0043] In the nucleic acid replication project of using existing nucleic acid molecules as templates and synthesizing new nucleic acid molecules by nucleic acid polymerase through the principle of complementary base pairing, the secondary structure of single-stranded nucleic acid molecules as templates will affect the efficiency of nucleic acid replication. When the structure is sufficiently stable, it may also lead to failure of nucleic acid replication. For example, in the PRC reaction, during the annealing stage of the reaction, the temperature drops rapidly, and the intramolecular secondary structure of the template is formed before the hybridization product of the primer and the template due to kinetic advantages. When the formed intramolecular secondary structure is stable enough , in the extension stage, it will hinder the extension of the primer by the nucleic acid polymerase, resulting in premature termination of the reaction or a reduction in reaction efficiency.
[...
Embodiment 2
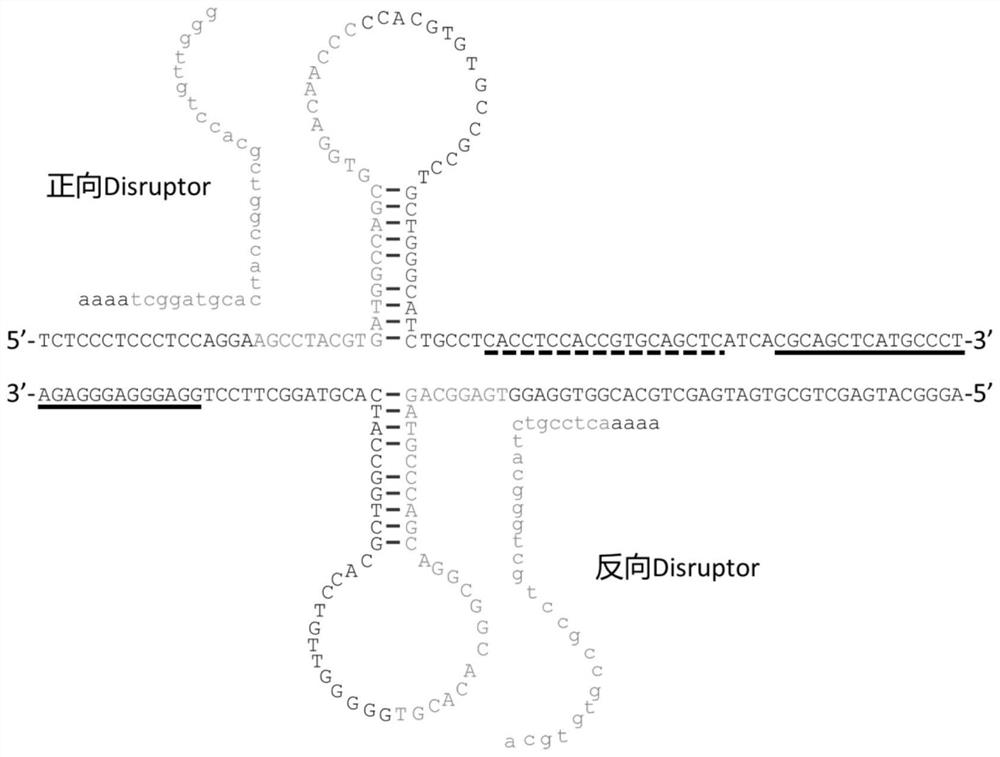
[0053] The present invention provides a method for duplicating nucleic acid fragments. In the in vitro replication reaction system of a single-stranded nucleic acid template with a secondary structure, the above-mentioned reagents for improving the replication efficiency of a single-stranded nucleic acid molecule with a secondary structure are added. The reagent destroys the base pairing in the secondary structure in the template, so as to reduce the hindering effect of the secondary structure in the molecule on the nucleic acid polymerase, that is, to prevent the secondary structure from hindering the extension of the nucleic acid polymerase to the primer during the extension stage, so as to Improve the efficiency of nucleic acid fragment replication.
[0054] Specifically, the method includes the steps of: adding a certain concentration of the reagent to the reaction system before the nucleic acid fragment replication reaction starts; the reaction initial concentration of the...
Embodiment 3
[0064] The present invention provides a reagent for improving the replication efficiency of a single-stranded nucleic acid molecule having a secondary structure as described above or the method for replicating a nucleic acid fragment as described above in a PCR reaction or a sequencing reaction or a transcription reaction or a reverse transcription reaction application. Specifically, the above-mentioned unzipping agent or the above-mentioned nucleic acid fragment can be applied to any molecular biology method that uses a single-stranded nucleic acid as a template and the template has a secondary structure, and synthesizes a new nucleic acid molecule by a nucleic acid polymerase through the principle of complementary base pairing In the replication method, the secondary structure in the template is melted by a melting agent, so that the above-mentioned molecular biology method can be carried out smoothly, so as not to affect the progress of the extension stage due to the stabili...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com