Application of rice transcription factor OsWRKY53 in negative regulation and control of cold tolerance of rice in booting stage
A technology of rice transcription factor and transcription factor, which is applied in application, genetic engineering, plant gene improvement, etc., can solve problems such as poor differentiation of branches and spikelets, decreased seed setting rate, and reduced number of grains per panicle, so as to improve rice booting. The effect of long-term cold resistance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Example 1: Biological function verification of rice booting stage negative regulator OsWRKY53
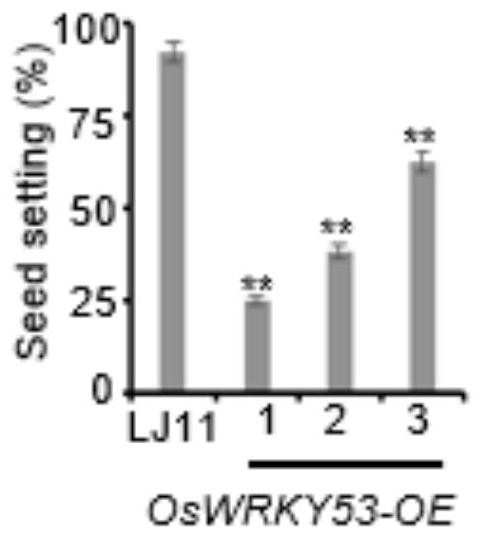
[0039] Observation of wild rice variety Longjing 11, oswrky53 mutant plants and OsWRKY53 overexpression transgenic plants under the background of Longjing 11 found that the seed setting rate of OsWRKY53 overexpressing plants was significantly lower than that of the control material Longjing 11 (such as figure 1 , 2 shown). Compared with the control material Longjing 11, the seed setting rate of the oswrky53 mutant plants was basically the same (such as image 3 , 4 shown). This suggests that OsWRKY53 is likely to be involved in the regulation of cold tolerance at the booting stage of rice.
[0040] Using wild rice varieties Longjing 11, oswrky53 mutant plants and overexpression transgenic plants under the background of Longjing 11 as experimental materials, the distance between the flag leaf and the second leaf of the rice was -5 ~ 0cm ( That is, when the anthers are in ...
Embodiment 2
[0043] Example 2: Detection of GA content in rice anthers negatively regulated by OsWRKY53
[0044] (1) Determination of GA content in anthers of Longjing 11 and oswrky53 mutant plants under normal and low temperature treatment
[0045] The wild rice variety Longjing 11 and the oswrky53 mutant plants under its genetic background were used as experimental materials. The distance between the flag leaf and the second leaf of the rice was -8 ~ -6cm (that is, the anthers were in the meiosis state). During the division stage), the rice was treated at 18°C for 15 days (the anthers were in the three-nucleated stage), and the anthers of about 30 rice main tillers were sampled, and the GA content in the anthers was detected by Thermo Scientific Ultimate 3000 UHPLC and TSQ Quantiva in Wuhan Lvjian Company .
[0046] The result is as Figure 10 As shown, □ means LJ11-NC, ■ means oswrky53-NC, Indicates LJ11-LT, Indicates oswrky53-LT. Low temperature stress will lead to the decrease ...
Embodiment 3
[0063] Example 3: Verification that OsWRKY53 directly binds to and inhibits the transcriptional activity of GA-related biosynthetic genes:
[0064] (1) Sequence analysis of GA-related biosynthetic genes
[0065] According to the 2000bp sequence of the OsGA20ox1, OsGA20ox3 and OsGA3ox1 promoter regions published in the Rice Genome Annotation Project, it was found that there were 2 W-boxes (TTGACC) in the 2000bp promoter region of the OsGA20ox1 gene and 1 W-box in the 2000bp promoter region of the OsGA20ox3 gene (TTGACC), there is a W-box (TTGACC) in the 2000bp promoter region of OsGA3ox1 gene such as Figure 12 shown.
[0066] (2) EMSA experiments verified that OsWRKY53 directly binds to GA-related biosynthetic genes
[0067] 1. Purify the OsWRKY53 protein with the Mbp tag, transform the plasmid into Escherichia coli BL21, pick a single clone and put it in 3ml of liquid LB medium containing kana antibiotics, shake overnight at 37°C. Put the activated bacterial solution into ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com