Method for rapidly and quantitatively detecting capping efficiency of RNA
A technology for RNA capping and quantitative detection, which is applied in the field of RNA synthesis research and can solve problems such as complex operations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
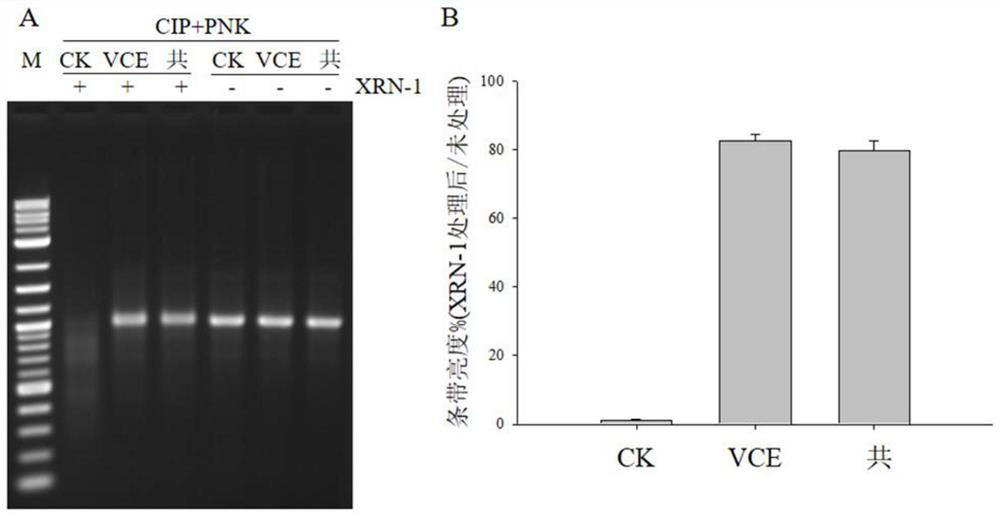
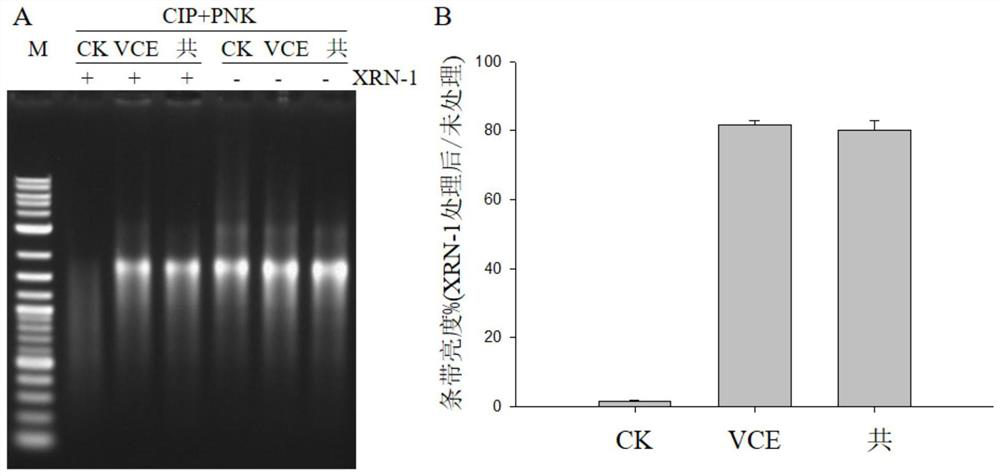
[0030] In this example, vaccinia virus capping enzyme and co-transcription method were used to cap sox7 RNA respectively, and then the calf intestinal alkaline phosphatase (CIP, #M0290) produced by NEB was used to dephosphorylate the RNA, and the RNA After purification, use T4 polynucleotide kinase (PNK, #M0201) produced by NEB to add monophosphate to the 5' end of RNA to obtain monophosphate RNA, and then use monophosphatase produced by NEB (XRN-1, #M0338 ) to digest monophosphorylated RNA, and finally run the gel to detect and quantitatively measure the brightness of the band, thereby distinguishing capped and uncapped RNA, and achieving the purpose of rapid quantitative detection of RNA capping efficiency. The specific steps are as follows:
[0031] S1. Synthesis of uncapped RNA by in vitro transcription: Using purified linearized DNA as a transcription template, KP34 RNA polymerase binds to a specific promoter on the template and initiates in vitro transcription of RNA to o...
Embodiment 2
[0057] The differences between this example and Example 1 are: (1) In this example, cas9 RNA is used for the reaction. (2) In steps S1 and S2 of Example 1, the KP34 RNA polymerase in the transcription and co-transcription reaction system was replaced with VSW3 RNA polymerase, and the transcription reaction was carried out at 16° C., and the transcription time was 16 hours.
[0058] The results of final gel running results and RNA capping efficiency are shown in the attached figure 2 shown.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com