Antibiotic drug resistance gene influence factor and difference analysis method
A technology of drug resistance genes and influencing factors, applied in the field of metagenomics bioinformatics analysis, can solve the problems of poor persuasiveness of differential antibiotic resistance genes and limited ability to annotate antibiotic resistance genes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0037] The present invention will be further described by the following examples, but these examples should not be used to explain the limitation of the present invention.
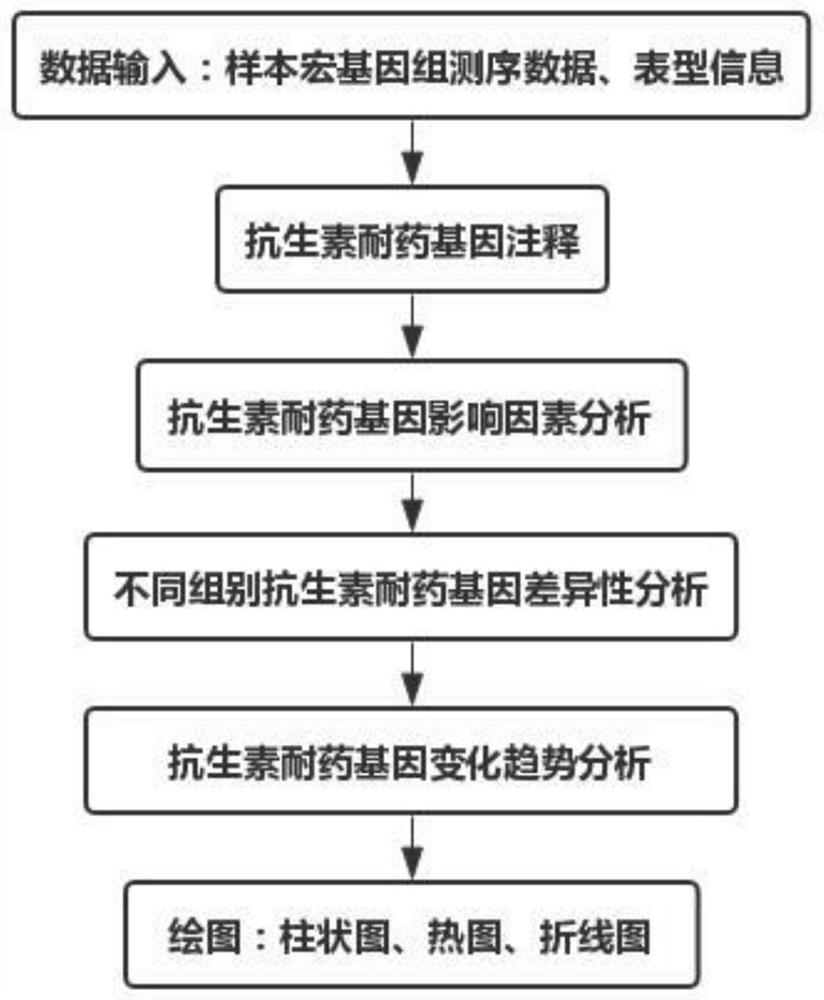
[0038] refer to figure 1 , the embodiment of the present invention is as follows:
[0039] 1. Data entry:
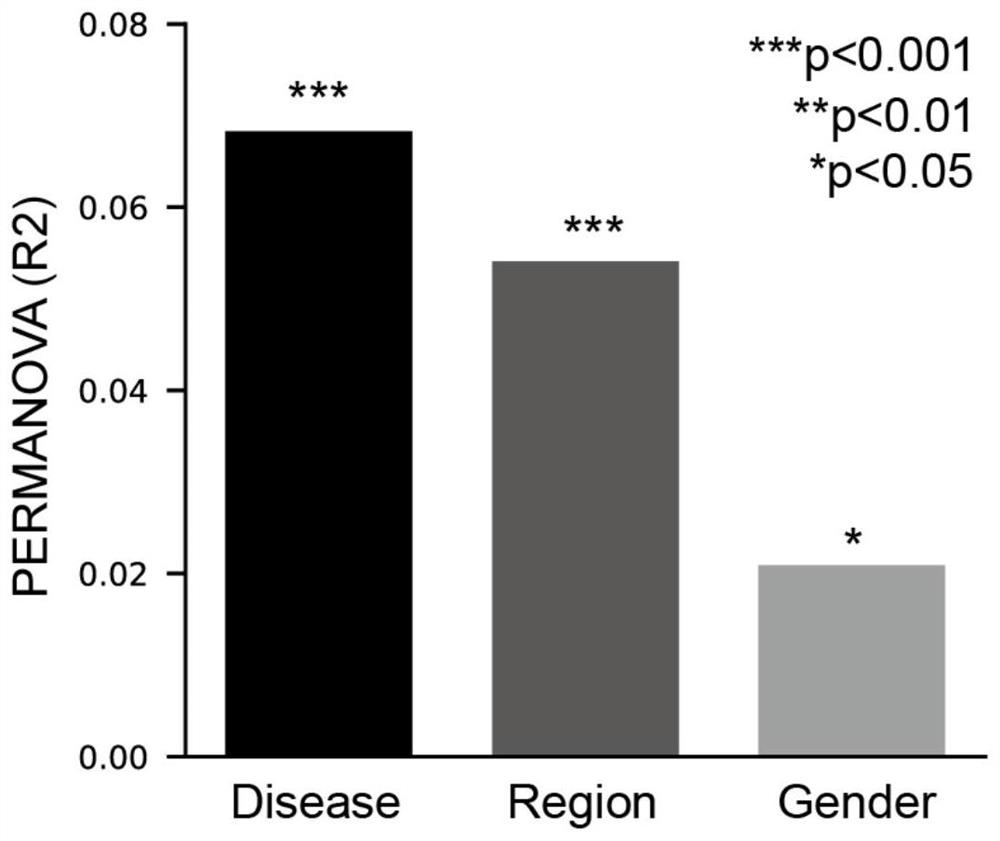
[0040] Specifically, the input data includes two parts: the first part is a txt file (sample_information.txt) separated by tabs, the first column and each row contain the sample ID, and the subsequent columns are the corresponding gender, disease status, Region and other phenotype and storage path information; the second part is the sample metagenomic sequencing data file, and the file name and storage path of each sample must be the same as the sample ID and storage path of the first part.
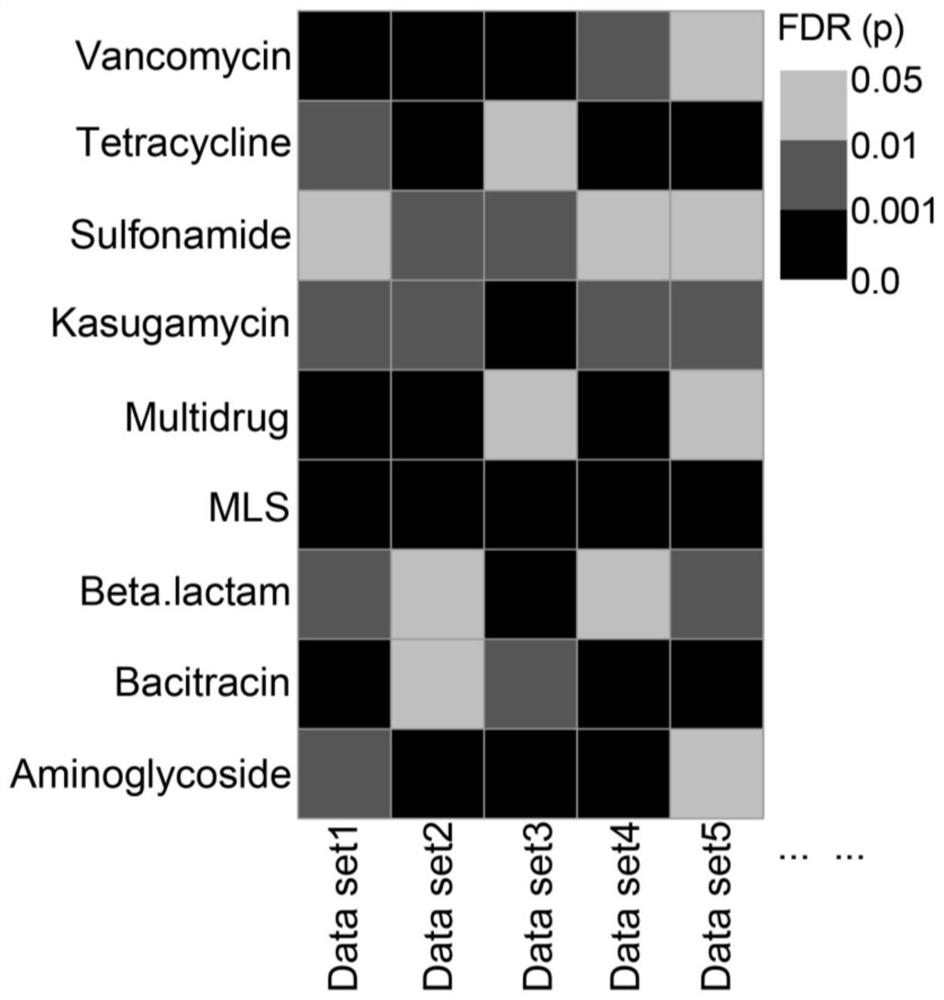
[0041] 2. Annotation of antibiotic resistance genes:
[0042] Specifically, based on the antibiotic resistance gene annotation tool ARGs-OAP and the database SARG v2.0, execute on the linux terminal: sh antib...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com