Primers for rapidly detecting fragmented new coronavirus S gene point mutation and application
A fragmentation and point mutation technology, applied in the fields of biochemistry and molecular biology, can solve problems such as difficult detection and severe fragmentation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Detect the new coronavirus S gene template with a length of only 50nt.
[0031] 1. Operation steps:
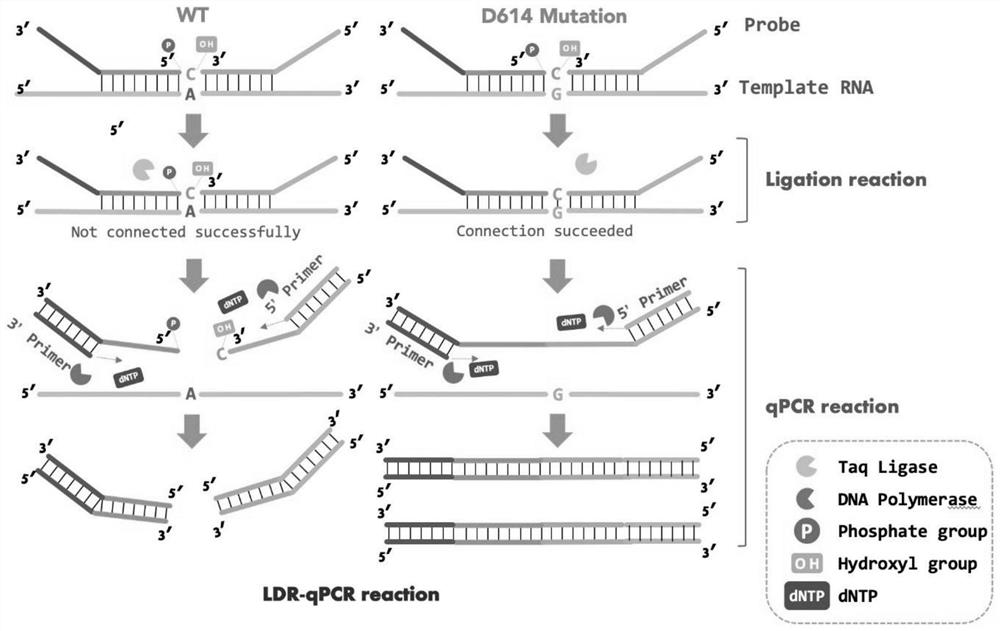
[0032] (1) The recombinant plasmid containing the D614G mutation target fragment of the new coronavirus S gene was transcribed in vitro to obtain a 50nt RNA fragment containing the D614G point mutation, which was used to simulate fragmented viral RNA.
[0033] (2) Use the RNA template obtained in (1) and the primer pair LDR-1 / LDR2 to prepare an LDR reaction mixture; set the 50nt wild-type S gene RNA obtained by the same method as in (1) as a control, and perform LDR reactions respectively. LDR reaction mixture includes: thermostable DNA ligase, RNA template, LDR primer pair, 10X reaction buffer and deionized water. The addition amount of each component in the LDR reaction mixture is shown in Table 2. Then start the LDR reaction, the reaction program is: ① 95°C for 3 minutes, ② 95°C for 30 seconds, ③ 58°C for 4 minutes. Steps ② and ③ were repeated for 35 cycles, and t...
Embodiment 2
[0042] The template length is smaller than the primer set, and the mutation can still be detected:
[0043] 1. Operation steps:
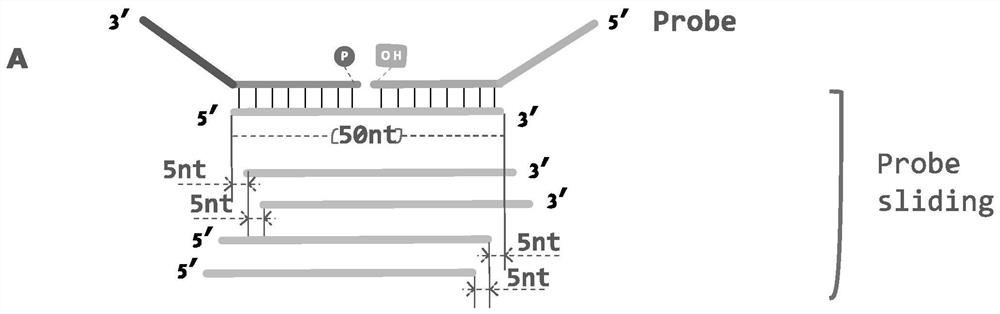
[0044] (1) Use different primers to transcribe the recombinant plasmid containing the target fragment of the new coronavirus S gene in vitro, and obtain different RNA fragments containing the D614G point mutation with a length of 50 nt, simulating the fragmented viral RNA that does not completely match the probe region of the LDR primer ( figure 2 ).
[0045] (2) Using the RNA obtained in (1) as a template, prepare an LDR reaction mixture with the primer pair LDR-1 / LDR2, and use 50nt wild-type S gene RNA as a control to perform LDR reactions respectively. The LDR reaction system and procedures are the same as Table 2 and Table 3.
[0046] (3) Using primers LDR-UF and LDR-UR to perform fluorescence quantitative PCR reaction detection on the reaction product of step (2).
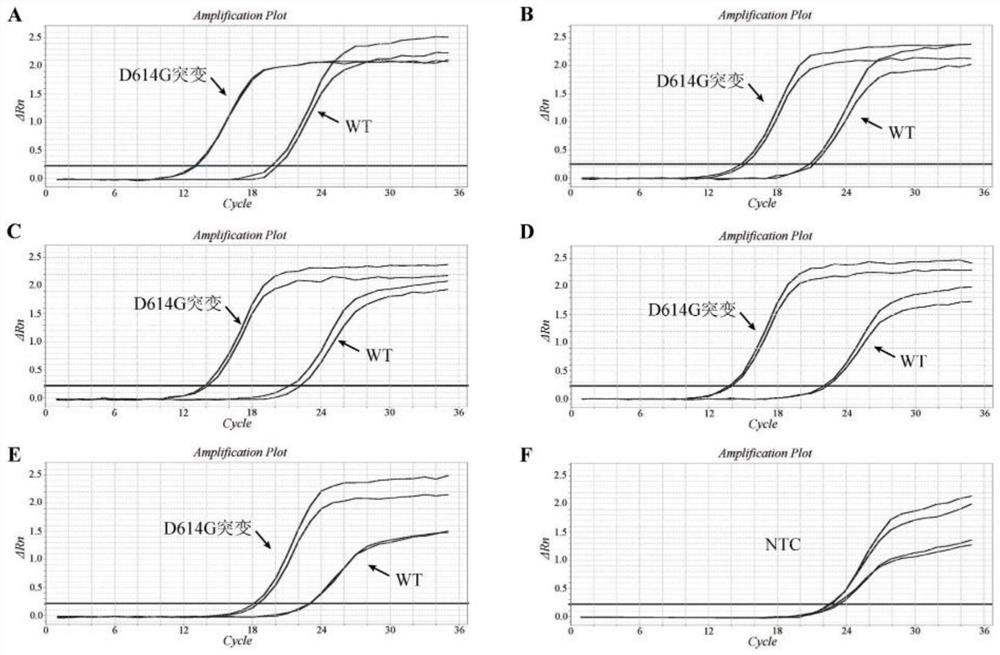
[0047] 2. Result analysis:
[0048] The experimental results are attached...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com