Ligand compound rapid pre-screening model based on deep learning
A ligand compound and deep learning technology, which is applied in the field of rapid pre-screening model of ligand compounds based on deep learning, can solve the problems of low affinity prediction accuracy, different performance, and success rate.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
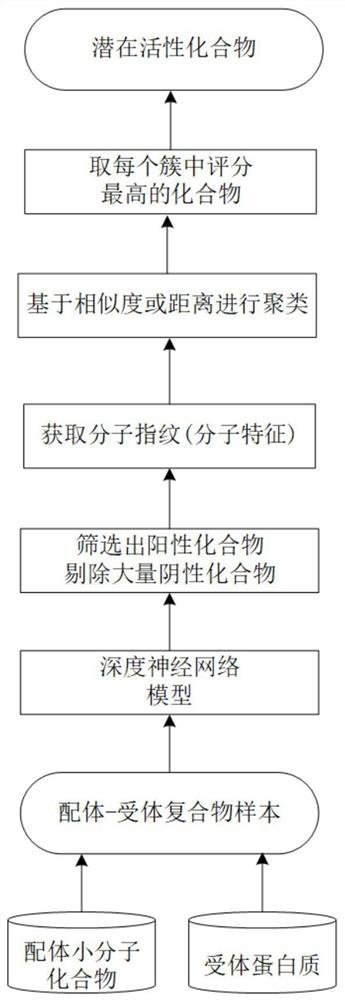
[0035] The following will be combined with figure 1 , clearly and completely describe the technical solution of the present invention,
[0036] Specifically, the present invention provides a rapid pre-screening model for ligand compounds based on deep learning, such as figure 1 As shown, it includes the following steps:
[0037] S1. Obtain the ligand small molecule compound through the DrugSpaceX database or ZINC database and the small molecule data set generated by the generated model, and obtain the receptor protein structure through the RCSB PDB database and PDBbind database;
[0038] S2. Use RDKit to generate SMILES strings of compound molecules, and use word2vec or similar algorithms to encode the compound molecules;
[0039] S3. Extract the amino acid sequence information of the receptor protein binding site (pocket), and encode it using word2vec or similar algorithms;
[0040] S4. Set the docking space range, combine the ligand and the receptor structure vector by a ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com