Fusion protein, base editing tool and application thereof
A fusion protein and gene technology, applied in the field of gene editing, can solve the problems of low targeting efficiency of editor PAM-preferred partial sites, which is not conducive to clinical research and application, etc., achieve good gene therapy prospects, broaden the targeting range and Scope of application, the effect of broadening the scope of targeting
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
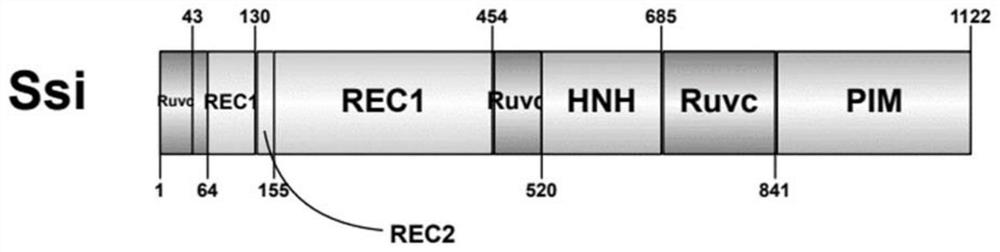
[0080] The amino acid sequence of the Cas9 protein homologue SsiCas9 derived from Streptococcus sinensis was compared with SpCas9, and the functional domains of SsiCas9 were divided. The domains are as follows: figure 1 shown, and find out the RuvC domain functional site of SsiCas9 (aspartic acid D9 at position 9) and mutate it into alanine (A), thereby obtaining SsiCas9 D9A nickase, its amino acid sequence is as SEQ ID NO. 1.
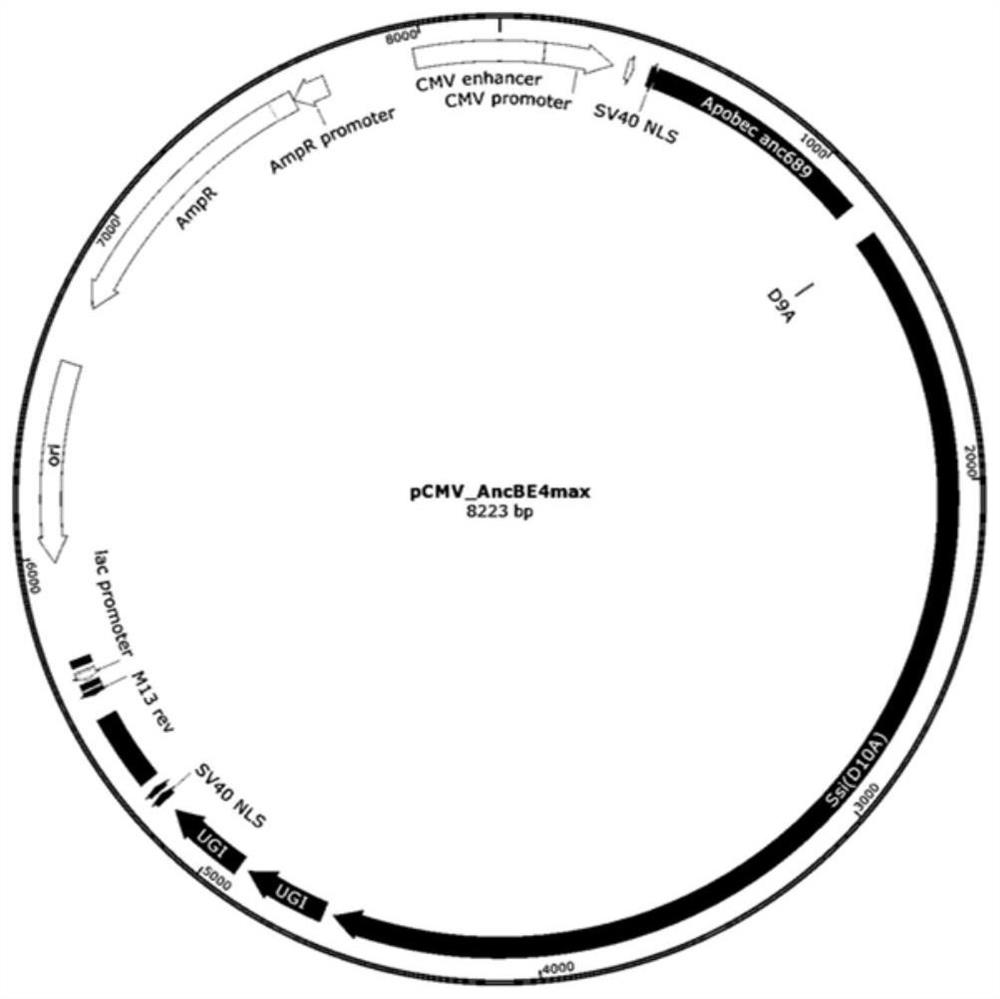
[0081] The prokaryotic codon of SsiCas9 D9A of Streptococcus sinensis was optimized for eukaryotic expression, so as to obtain the coding DNA sequence of SsiCas9 D9A suitable for expression in eukaryotic cells, as shown in SEQ ID NO.2. After optimization, the whole gene synthesis of SsiCas9 D9A commercial company. The construction strategy is to replace the SpCas9 D10A of ancBE4max with SsiCas9D9A on the basis of ancBE4max, where ancBE4max is synthesized by a commercial company. In the next step, we excised part of the XTENlinker-SpCas9 D10A-10aa lin...
Embodiment 2
[0090] 2.1 Vector Construction of SsiCas9-ancBE4max System gRNA Plasmid
[0091] Using pGL3-U6-sgRNA (Addgene #51133) as the expression backbone, a gRNA expression vector suitable for the SsiCas9 gRNA editing system was constructed. According to the tandem repeat sequence derived from Streptococcus sinensis, the scaffold sequence suitable for the SsiCas9 gRNA interaction system was designed, and the scaffold (suitable for SpCas9) of pGL3-U6-sgRNA (Addgene #51133) was replaced with the SsiCas9 gRNA scaffold (suitable for SsiCas9), The successfully constructed complete plasmid is shown in SEQ ID NO.7, named pGL3-U6-Ssi gRNA, and its plasmid structure is shown in Figure 4 . The restriction sites connected to the targeting gRNA sequence are two BsaI restriction sites, and the plasmid is synthesized by a commercial company.
[0092] 2.2 Construction of SsiCas9-ancBE4max system targeting gRNA plasmid
[0093] Design gRNA and synthesize two complementary paired oligos, the upstre...
Embodiment 3
[0099] The base editing system composed of the SsiCas9-ancBE4max plasmid and the pGL3-U6-Ssi gRNA plasmid constructed in the above example was used to transfect HEK293T cells, and the process was as follows:
[0100] 3.1 HEK293T cells (from ATCC) were revived and cultured in a 10 cm culture dish (Corning, 430167). The medium was DMEM (HyClone, SH30243.01) mixed with 10% fetal bovine serum (HyClone, SV30087). The culture temperature was 37°C, and the carbon dioxide concentration was 5%. After multiple passages, when the cell density reached 90%, the cells were divided into 24-well plates.
[0101] 3.2 Observe the state of the cells after three passages of HEK293T cells have been resuscitated. Plate the cells in good condition in a 24-well plate. After the plated cells are cultured for 18-24 hours, transfect them when the cell concentration is 80%. The dosage of each component during the transfection process : SsiCas9-ancBE4max plasmid: 1 μg, pGL3-U6-Ssi gRNA plasmid: 0.5 μg, E...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com