Fusion protein and base editing tool and method and application thereof
A fusion protein and base technology, applied in the field of gene editing, can solve problems such as limiting the scope of genome targeting, and achieve the effect of broadening the scope of targeting
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0109] First construct ancBE4max-NG and ABEmax-NG plasmids, and introduce 7 amino acid mutations (R1335V / L1111R / D1135V / G1218R / E1219F / A1322R / T1337R) into ancBE4max and ABEmax plasmids through Mut Express II FastMutagenesis Kit V2 (Vazyme, C214-02) , ancBE4max was synthesized from the whole gene by a commercial company, and the ABEmax plasmid was purchased from Addgene (#112095). The DNA sequence contained in the generated pCMV-ancBE4max-NG is shown in SEQ ID No.17; the DNA sequence contained in pCMV-ABEmax-NG is shown in SEQ ID No.18.
Embodiment 2
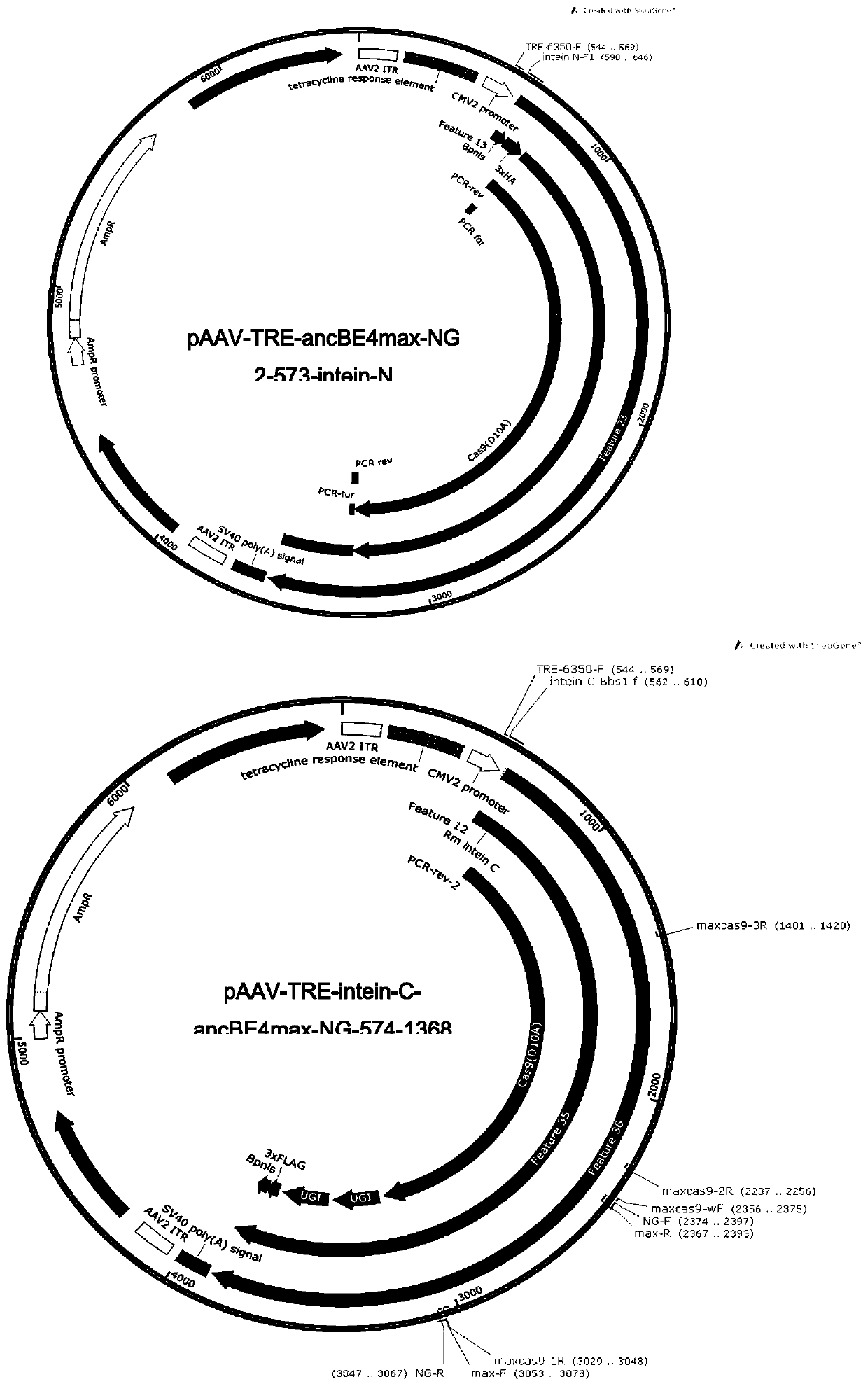
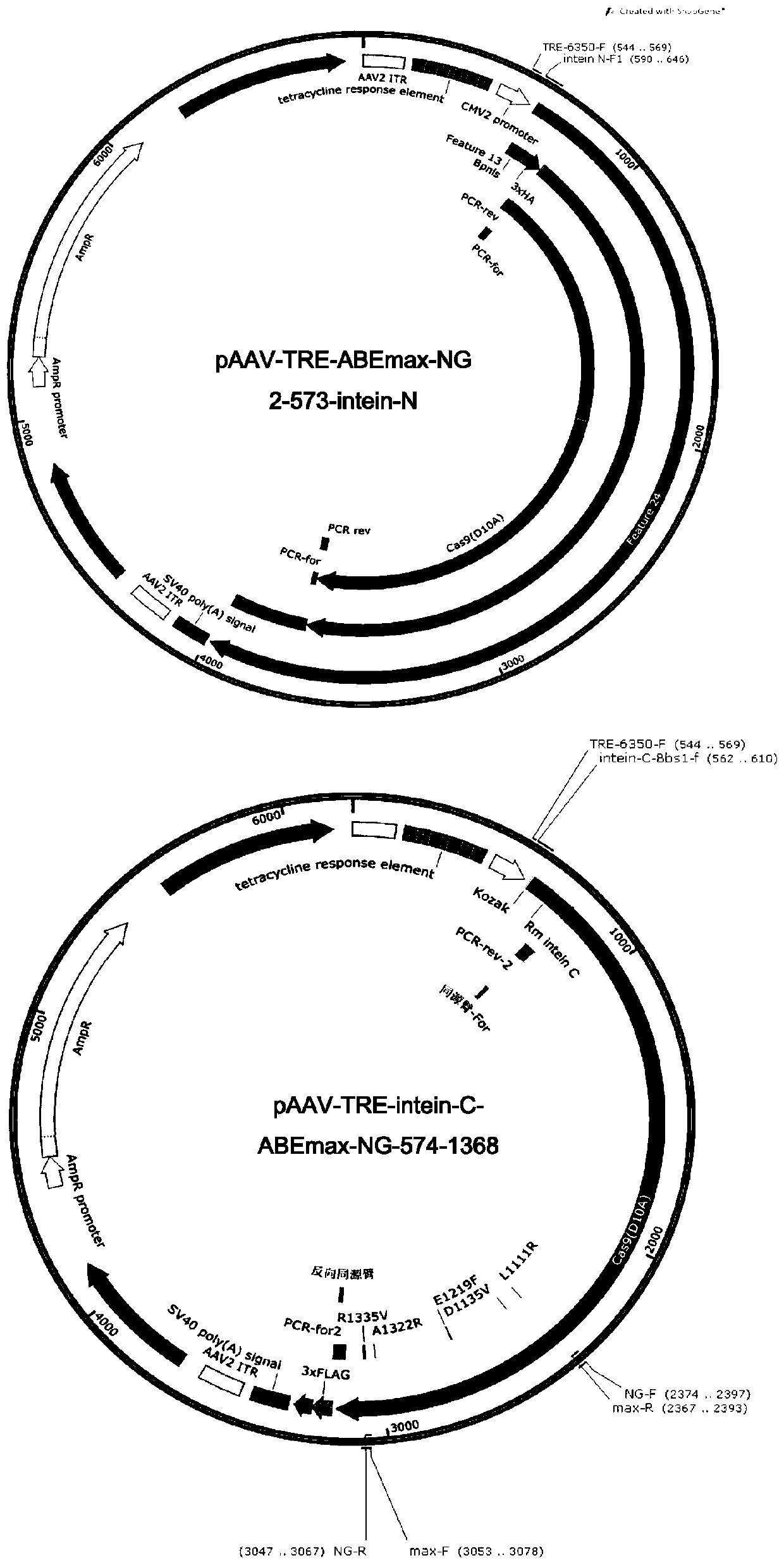
[0111] On the basis of embodiment 1 gained ancBE4max-NG and ABEmax-NG, construct as figure 1 and 2 Indicated pAAV-TRE-ancBE4max-NG 2-573-intein-N, pAAV-TRE-intein C-ancBE4max-NG 574-1368, pAAV-TRE-ABEmax-NG 2-573-intein-N, pAAV-TRE -intein C-ABEmax-NG 574-1368.
[0112] 2.1 pAAV-TRE-ancBE4max-NG 2-573-intein-N, pAAV-TRE-intein C-ancBE4max-NG 574-1368, pAAV-TRE-ABEmax-NG 2-573-intein-N, pAAV-TRE-intein Construction of C-ABEmax-NG574-1368 plasmid
[0113] The PCR primers whose sequences are shown in Table 1 were synthesized by Jinweizhi Biotechnology Co., Ltd., diluted to 10 μM as PCR primers, and the original pAAV-TRE was used as a template.
[0114] Table 1
[0115]
[0116]
[0117] The Novizym high-fidelity enzyme kit (Vazyme, p501-d2) was used to amplify the vector sequence fragment and the N-terminal or C-terminal fragment of ABEmax or ancBEmax, respectively. Amplification system (see Table 2) and PCR reaction conditions are as shown:
[0118] Table 2
[0119]...
Embodiment 3
[0128] Using the N-ancBE4max-NG+C-anc-BE4max-NG and N-ABEmax-NG+C-ABEmax-NG systems constructed in the above examples to transfect HEK293T cells, the process is as follows:
[0129] 3.1 HEK293T cells (from ATCC) were revived and cultured in a 10 cm culture dish (Corning, 430167). The medium was DMEM (HyClone, SH30243.01) mixed with 10% fetal bovine serum (HyClone, SV30087). The culture temperature was 37°C, and the carbon dioxide concentration was 5%. After multiple passages, when the cell density was 80%, the cells were divided into 12-well plates. The 12-well plate was coated with a 1:10 diluted polylysine solution (Sigma, P4707-50ML) before use.
[0130] 3.2 When the cell concentration is 80%, replace the medium with DMEM medium with 10% serum, and culture for 2 hours to restore the best cell state. The amount of plasmid transfected in each hole is N-ancBE4max-NG 0.5ug, C-anc-BE4max-NG 0.5ug, sgRNA0.5ug or N-ABEmax-NG 0.5ug, C-ABEmax-NG 0.5ug, sgRNA0. 5ug was co-transfec...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com