Coding gene sequence with adenosine triphosphate tunneling metalloenzyme activity
A technology of adenosine triphosphate tunneling and encoding gene, which is applied in the field of adenosine triphosphate tunneling metalloenzyme, and can solve the problem of insufficient understanding of TTM protein and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0019] Amplification of the ZjAC1 gene
[0020] The RNA of winter jujube fruit was extracted, and then reverse-transcribed into cDNA as a PCR amplification template for PCR amplification; wherein, the forward primer: 5'-ATGAGAGGGAAAAATCACAT-3', the reverse primer: 5'-TCAGCTAGGCAACTTACGGG-3'.
[0021] PCR amplification program: 94°C, 4min; (94°C, 40s; 55°C, 45s; 72°C, 50s) 35 cycles; 72°C, 10min.
[0022] The PCR products were subjected to agarose gel electrophoresis to observe the amplification results, and the candidate target bands were recovered by agarose gel, and the recovered products were connected into the pMDTM19-T cloning vector for sequencing to obtain the ZjAC1 sequence as a candidate date adenylate cyclization Enzyme gene for functional verification.
Embodiment 2
[0024] coli-deficient complementation assay
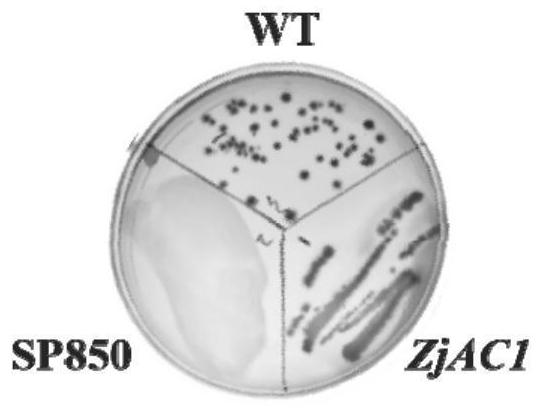
[0025] Cultivate Escherichia coli cyaA-deficient SP850 strain, SP850 strain lacks adenylyl cyclase gene (AC), transform the pET-15b recombinant plasmid with ZjAC1 gene into SP850 after heat shock, coat with ampicillin and kana antibiotics On a petri dish, pick a single clone and culture it until the OD600 is 0.5, then induce expression and culture it for 4 hours, culture it by streaking on MacConkey medium, and observe.
[0026] It was found that, if figure 1 , the color of the colonies of the SP850-deficient strain and the wild-type E. coli strain with the recombinant plasmid was red, while the color of the SP850-deficient strain was white in MacConkey medium, indicating that the ZjAC1 gene made up for the lack of cyaA in the SP850 strain gene, which functions as the adenylyl cyclase gene.
Embodiment 3
[0028] Protein Catalytic Test in Vitro
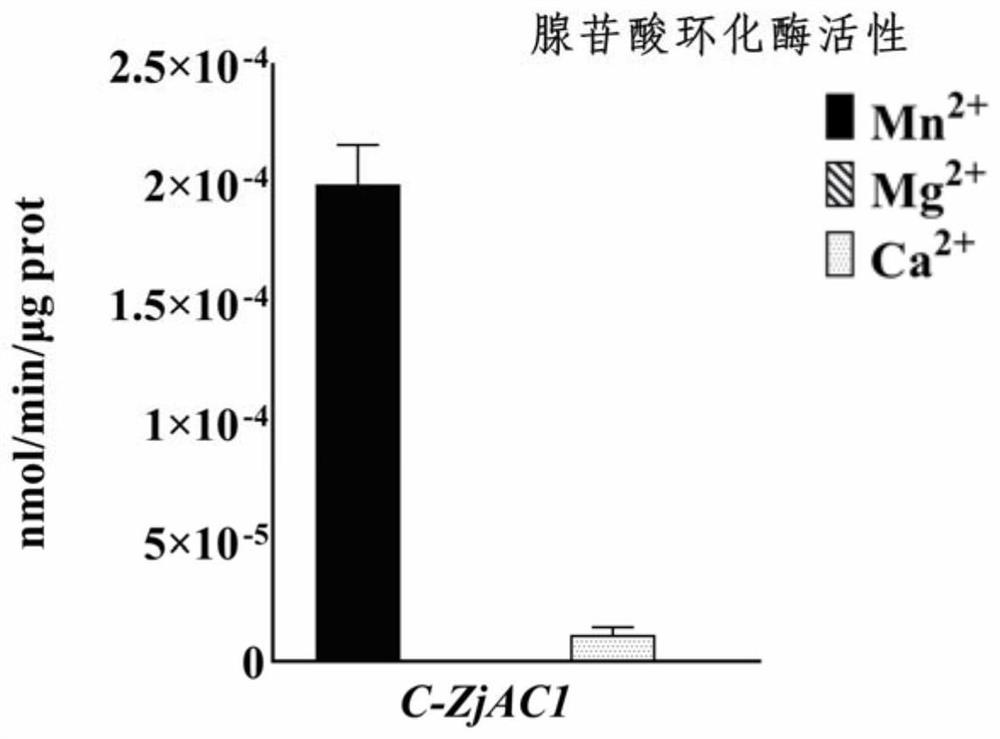
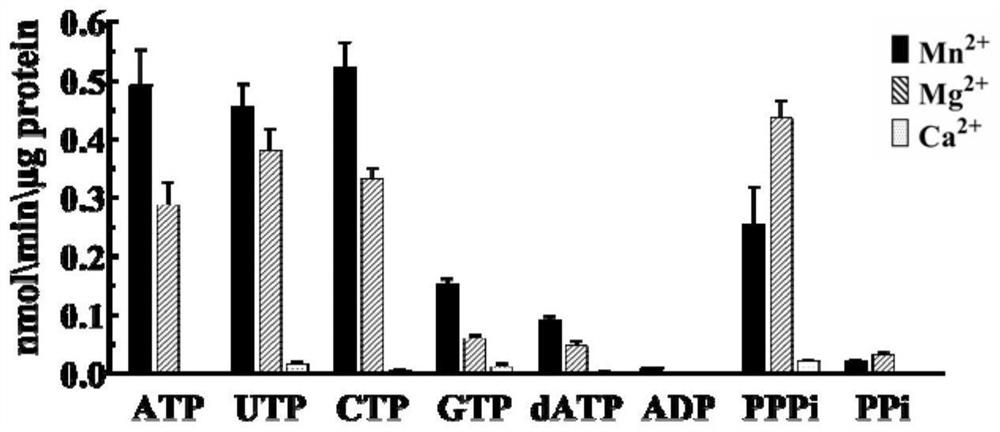
[0029] The pET-15b recombinant vector carrying the ZjAC1 gene was transformed into Escherichia coli BL21 strain, cultured at 37°C and 200rpm, and then the protein was extracted and purified, and the obtained protein was used for the detection of enzyme catalytic activity. The results were as follows: Figure 2-3 . It was found that when manganese ions were used as coenzyme ions, cAMP was produced in the catalytic product, which indicated that ZjAC1 protein could catalyze ATP to form cAMP in vitro, and played the role of AC.
[0030] AC enzyme activity detection method:
[0031] Catalytic system: 50mM TRIS-HCl (pH 7.5), 0.5mM ATP and 20mM coenzyme metal ion, the catalytic condition is 30°C, 20min. After the catalysis is completed, high-performance liquid chromatography (HPLC) is used to detect the cAMP content in the catalytic system, so as to determine the catalytic activity and efficiency of the enzyme.
[0032] The detection parame...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com