Nucleic acid ligase
A ligase and nucleic acid technology, applied in the biological field, can solve the problem that the ligase ligation efficiency is not high enough
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1. Construction of mutant HyperLigase protein expression plasmid by site-directed mutagenesis
[0031]Starting from the prior art HyperLigase (SEQ ID NO: 1), the applicant screened out potential mutation sites through analysis and research, namely Arg79, Lys249, Lys370 and Lys372 of HyperLigase.
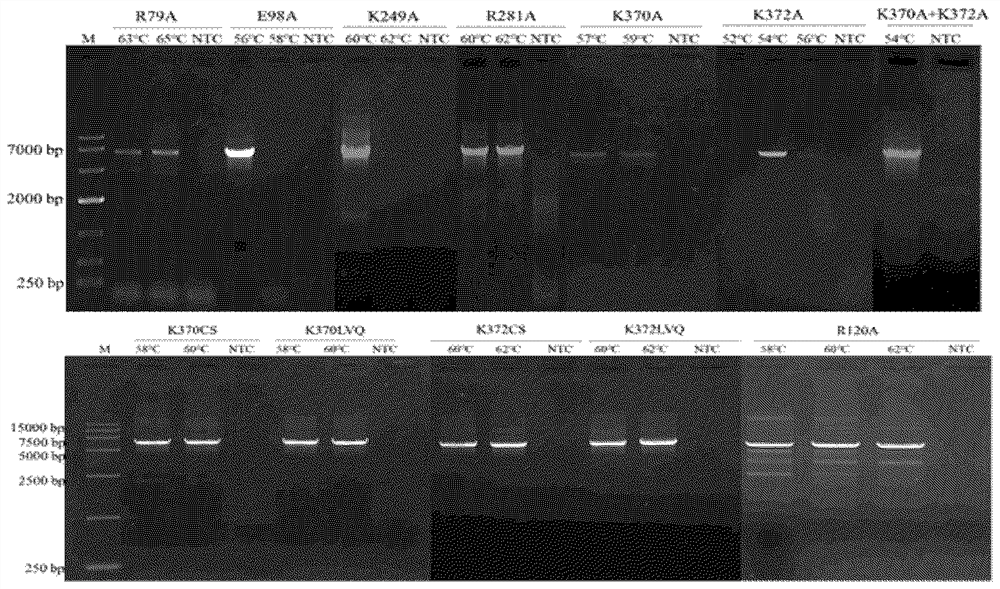
[0032] According to the screened mutation sites and the corresponding amino acids to be transformed, use the NEBasechanger site-directed mutagenesis primer online design tool (https: / / nebasechanger.neb.com) to design back-to-back point mutation primers. After synthesizing the corresponding primers, use the wild-type HyperLigase protein (SEQ ID NO: 1) expression plasmid as a template, and use high-fidelity DNA polymerase to carry out PCR reaction. figure 1 ). After purification, the DNA was phosphorylated at the end and circularized by self-ligation, and the product was transformed into BL21(DE3) competent cells, spread on a plate containing antibiotics, and the coloni...
Embodiment 2
[0034] Example 2. Protein induced expression and purification
[0035] Pick positive colonies from the plate and inoculate them into LB medium for resuscitation. After resuscitation, the bacterial solution is expanded by 10 to 50 times, and transferred to 1L LB medium for cultivation until the OD600 absorbance is 0.6 to 0.8, so that the bacterial solution reaches the logarithmic growth phase. , add an IPTG inducer to a final concentration of 0.1-1 mmol / L, and induce culture at 14-37°C for 6-24 hours. After collecting the bacterial cell precipitate, the bacterial cell was crushed by ultrasonic treatment, and the ultrasonic crushing product was purified. Purify using manual method or instrument method, according to the instrument and equipment guide, after elution, the concentration of the protein is measured and converted to molar concentration, and the purity is identified by SDS-PAGE (see image 3 ). The detected mutant HyperLigase was stored in 50% glycerol solution at -20...
Embodiment 3
[0037] Enzyme activity comparison of HyperLigase after mutation with original Hyperligase and CircLigase
[0038] CircLigase is currently the single-stranded DNA ligase with the best activity known on the market. In order to test the difference in activity between HyperLigase and CircLigase before and after mutation, a linear ligation reaction of intermolecular ligation was used to compare the activities of the two enzymes.
[0039] The CircLigase linear connection reaction system is as follows:
[0040]
[0041] The HyperLigase linear ligation reaction system is as follows:
[0042]
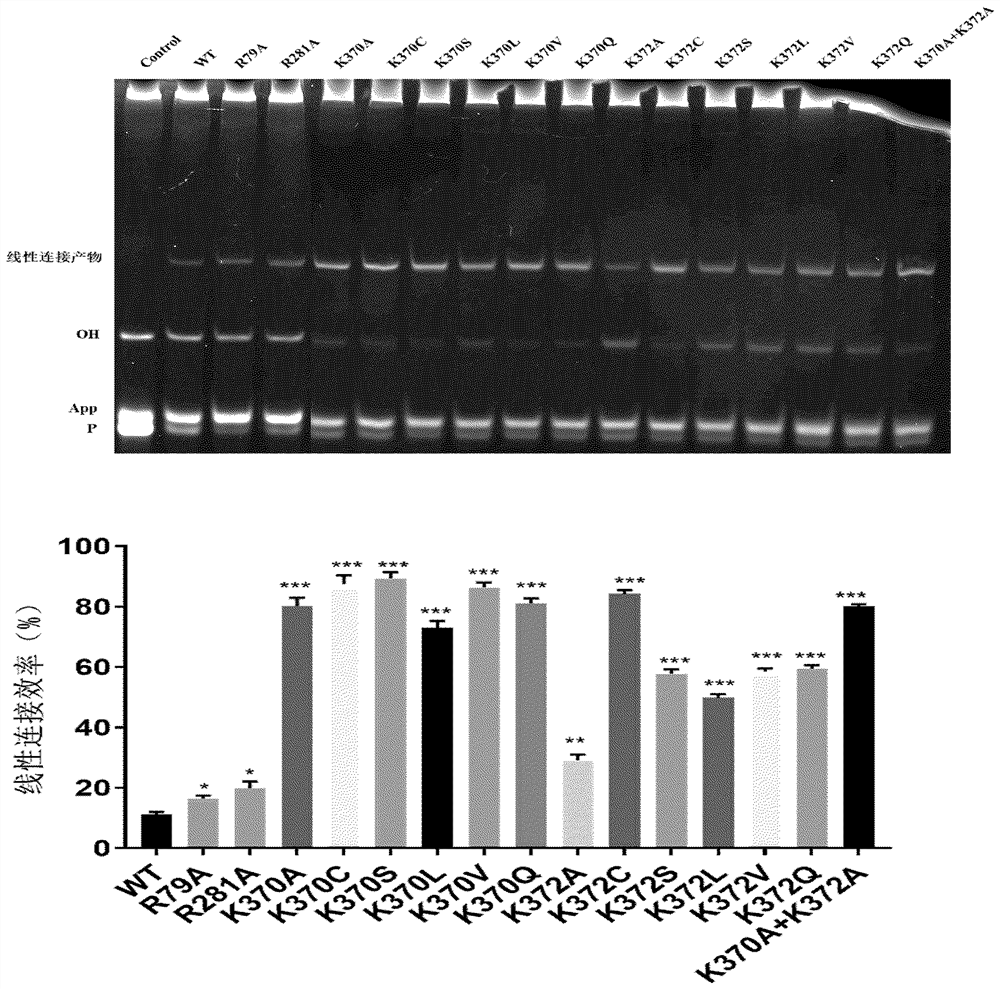
[0043] The HyperLigase reaction was carried out at 75°C, and the CingcLigase reaction was carried out at 60°C, and the reaction time was 6h. After the reaction was completed, Urea-PAGE electrophoresis identification and grayscale analysis compared the activity differences of the enzymes (see Figure 4 ).
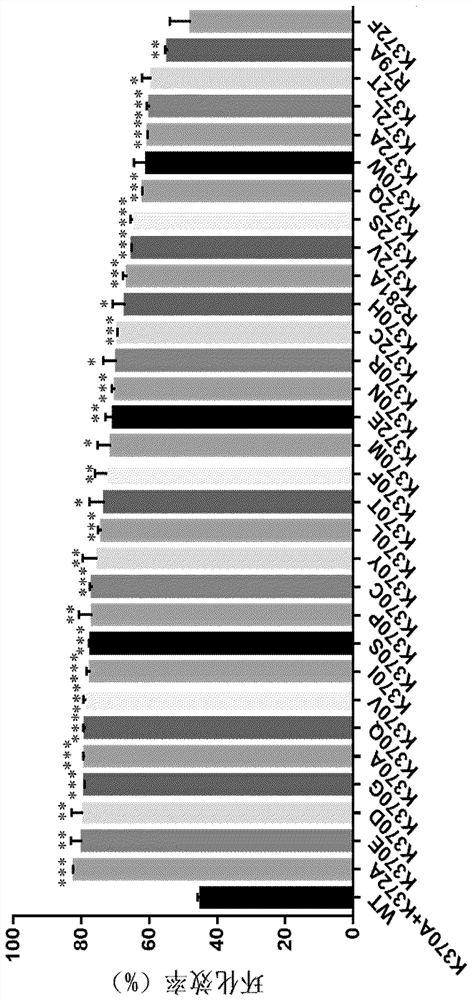
[0044] The test results showed that K370P mutation, K372E mutation, and K370C muta...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com