Detection method for capturing cfDNA5mC fragment
A detection method and fragment technology, which are used in the determination/inspection of microorganisms, biochemical equipment and methods, etc., to achieve high-performance results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
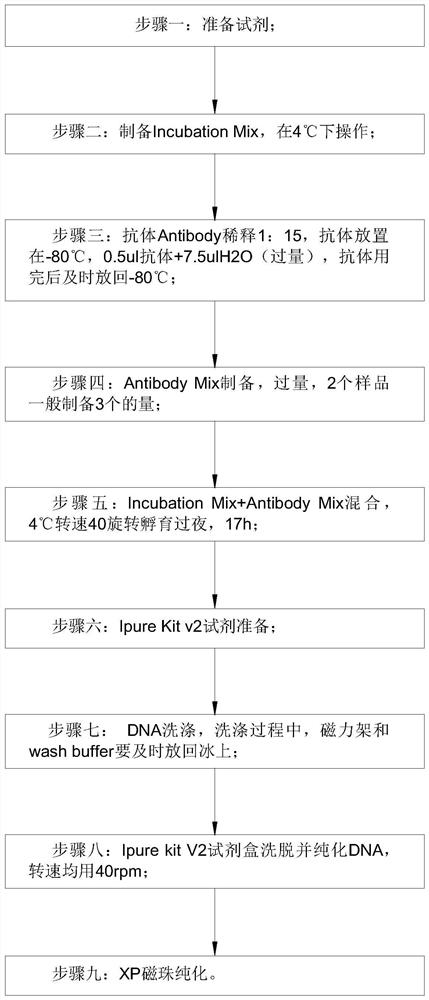
[0041] Embodiment one: if Figure 1-3 As shown, a detection method for capturing cfDNA5mC fragments of the present invention comprises the following steps:
[0042] Step 1: Prepare reagents;
[0043] Step 2: Prepare Incubation Mix and operate at 4°C;
[0044] Step 3: Dilute the antibody Antibody 1:15, place the antibody at -80°C, 0.5ul antibody + 7.5ulH2O (excess), put it back to -80°C in time after the antibody is used up;
[0045] Step 4: Antibody Mix preparation, excess, 2 samples generally prepare 3 quantities;
[0046] Step 5: Mix Incubation Mix+Antibody Mix, incubate overnight at 4°C and rotate at 40 for 17 hours;
[0047] Step 6: Ipure Kit v2 reagent preparation;
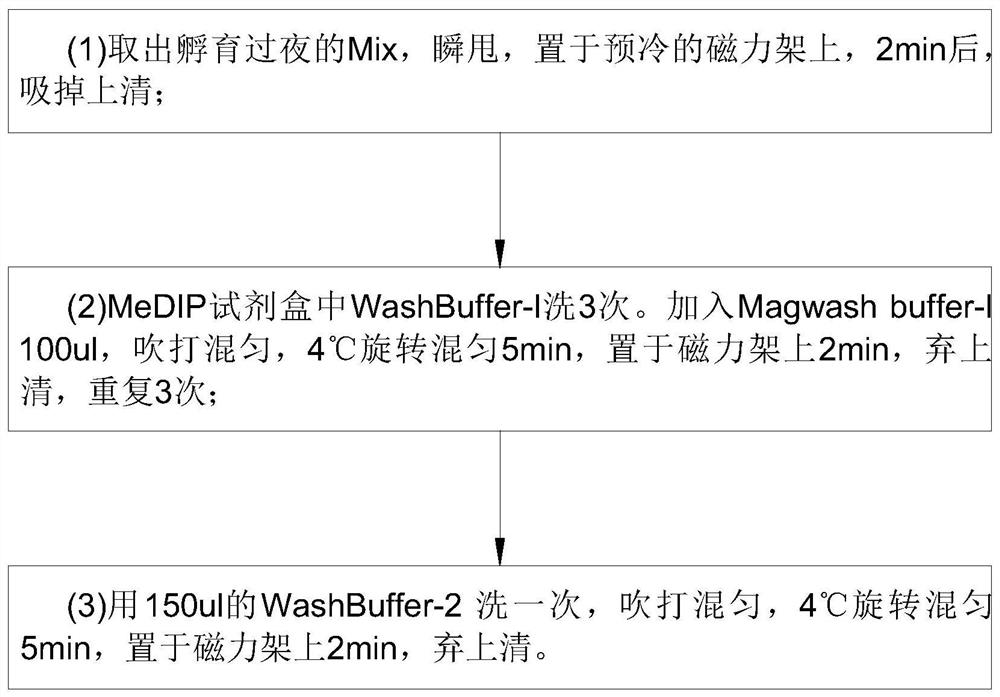
[0048] Step 7: DNA washing, during the washing process, the magnetic frame and wash buffer should be put back on ice in time;
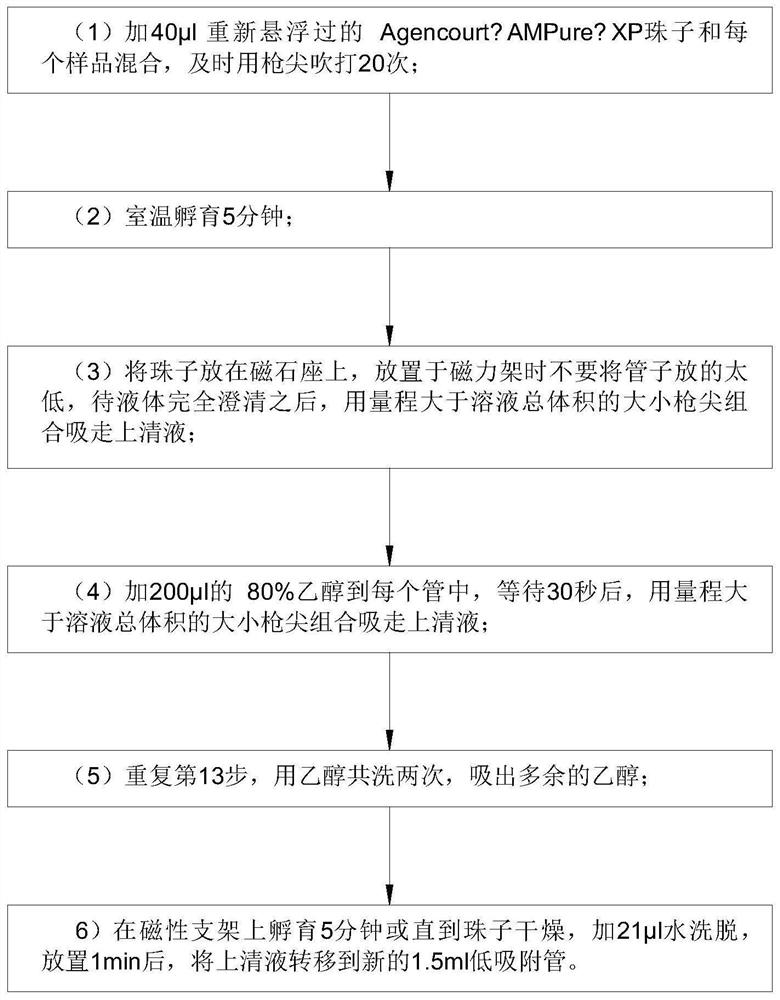
[0049] Step 8: Ipure kit V2 kit eluted and purified DNA at 40rpm;
[0050] Step 9: XP magnetic bead purification.
[0051] Wherein, in step 1, the reagent preparation process ...
Embodiment 2
[0072] Embodiment two: if figure 1 Shown, a detection method for capturing cfDNA5mC fragments, comprising the following steps:
[0073] Step 1: Prepare reagents;
[0074] Step 2: Prepare Incubation Mix and operate at 4°C;
[0075] Step 3: Dilute the antibody Antibody 1:15, place the antibody at -80°C, 0.5ul antibody + 7.5ulH2O (excess), put it back to -80°C in time after the antibody is used up;
[0076] Step 4: Antibody Mix preparation, excess, 2 samples generally prepare 3 quantities;
[0077] Step 5: Mix Incubation Mix+Antibody Mix, incubate overnight at 4°C and rotate at 40 for 17 hours;
[0078] Step 6: Ipure Kit v2 reagent preparation;
[0079] Step 7: DNA washing, during the washing process, the magnetic frame and wash buffer should be put back on ice in time;
[0080] Step 8: Ipure kit V2 kit eluted and purified DNA at 40rpm;
[0081] Step 9: XP magnetic bead purification.
[0082] Ipure kit V2 kit eluted and purified DNA (all speeds were 40rpm)
[0083] (1)DNA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com