Method for judging sample degradation based on CNV result
A sample and test sample technology, applied in genomics, instrumentation, proteomics, etc., can solve the problems that sample degradation or contamination cannot be judged, the result of judgment is inaccurate, and the judgment personnel have high requirements, so as to achieve high accuracy and easy operation Convenient and universal effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
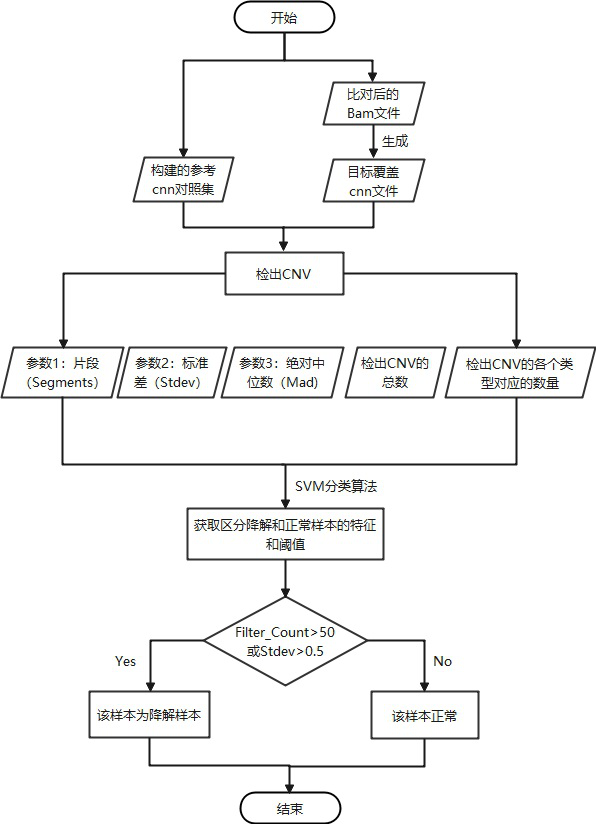
[0058] Please refer to figure 1 , the overall process is outlined as follows:
[0059] 1. Target coverage CNN file generation;
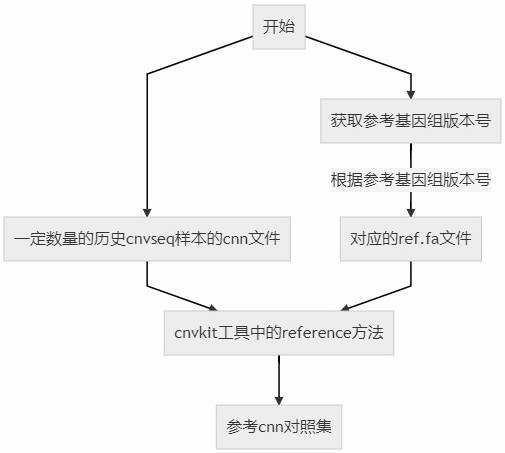
[0060] 2. Refer to the construction of the CNN comparison set;
[0061] 3. Detection of copy number variation;
[0062] 4. Acquisition of parameter indicators for automatic quality control;
[0063] 5. Acquisition of features and thresholds for distinguishing degraded and normal samples.
[0064] The operation steps of each part are described in detail below.
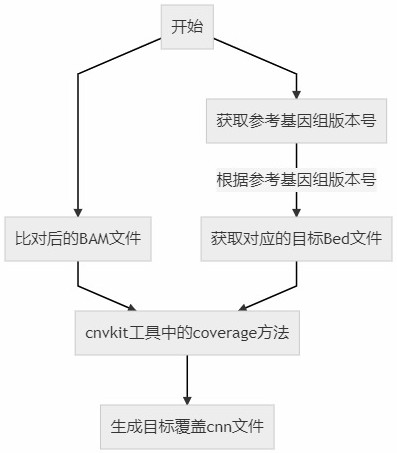
[0065] 1. Target coverage CNN file generation
[0066] According to the reference genome, the target coverage CNN file of the test sample is generated, which is mainly used to record the coverage of the compared bam file calculated according to the given area in the target bed file, and combined with the subsequent reference CNN control set (below) Named after Reference.cnn) for CNV detection.
[0067] Please refer to figure 2 , the build process is as follows:
[0068] The reference...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com