DNA methylation biomarker combination and application thereof
A methylation marker, methylation technology, applied in recombinant DNA technology, DNA/RNA fragment, determination/inspection of microorganisms, etc., to achieve the effect of speeding up diagnosis and treatment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
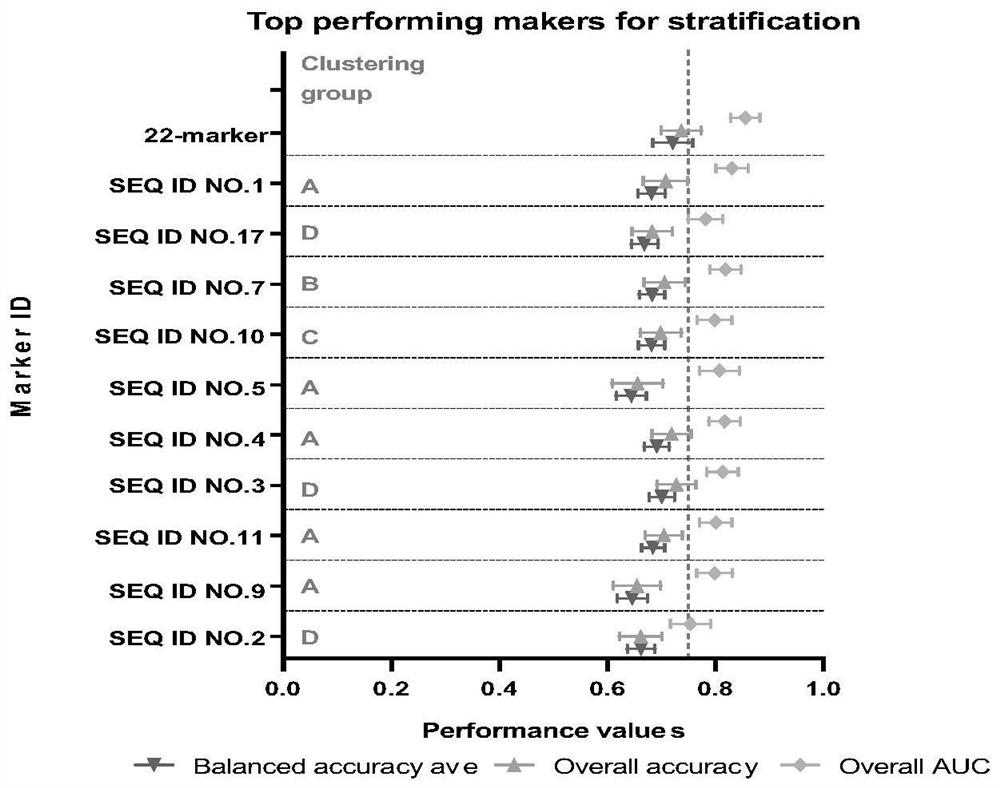
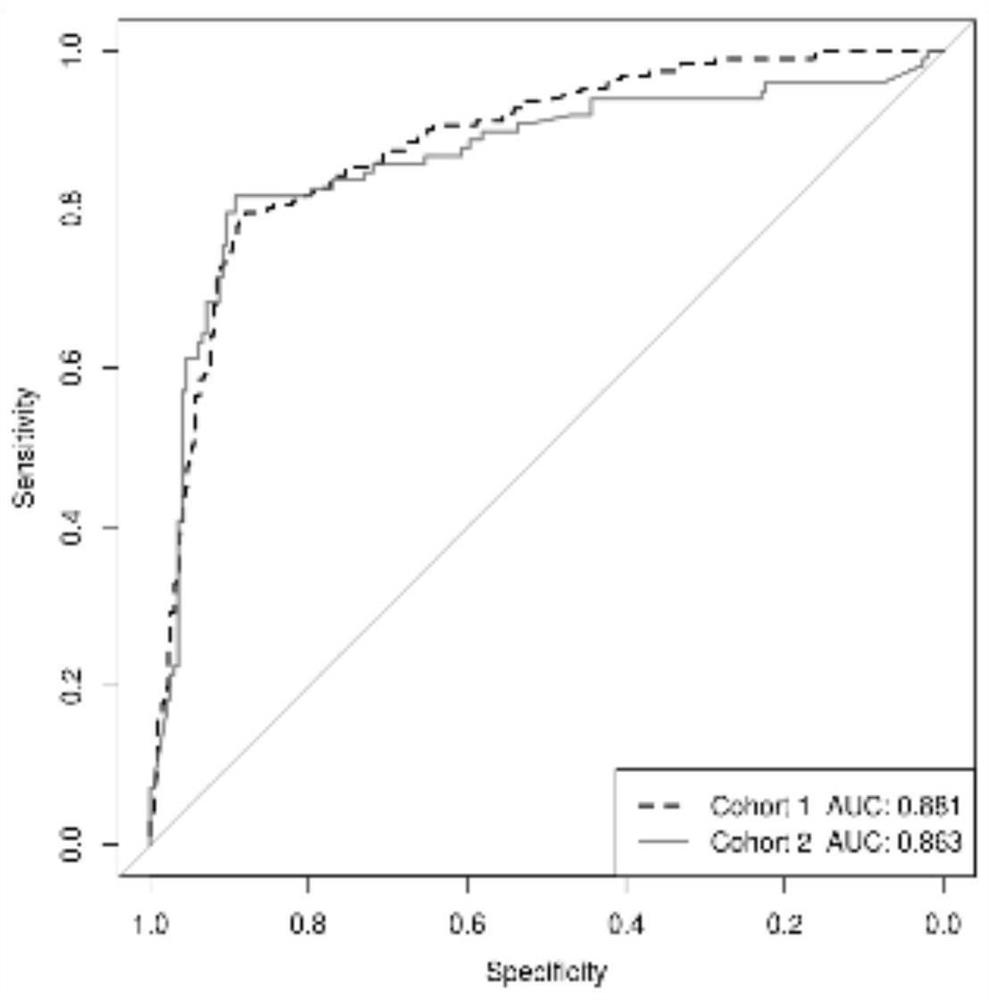
[0102] The sequence compositions including the 5 markers used for bladder cancer risk stratification correspond to the following table: SEQ ID NO.1, SEQ ID NO.2, SEQ ID NO.3, SEQ ID NO.5, SEQ ID NO.7 . In addition to the composition of the above-mentioned 5 markers, Table 1 also includes nucleic acid sequences of 22 other listed biomarkers. Each biomarker is indicated by the degree of co-methylation at multiple methylation sites indicated by the sequence [CG].
[0103] Table 1 Co-methylation composition of DNA methylated regions
[0104]
[0105]
[0106] For the above methylated sequences, specific primer pairs and probes for co-methylation of multiple methylated regions are shown in Table 2.
[0107] Table 2-1 Primer and probe sequence combination 1 for co-methylation detection of 22 methylated regions Table 2-1 Primer and probe sequence combination 1 for co-methylation detection of 22 methylated regions
[0108]
[0109]
[0110]
[0111] Table 2-2 Primer ...
Embodiment 2
[0123] Example 2 Multiplex fluorescent quantitative PCR for co-methylation detection of 5 methylated regions in 22 methylated regions
[0124] The 22 methylated regions comprising the 5 methylated biomarkers (SEQ ID NO.1-22) for co-methylation detection of every 2-3 methylated regions.
[0125] The specific process is as follows:
[0126] 1. DNA extraction
[0127] The extraction kit was purchased from QIAGEN, and was carried out according to the instructions of the kit.
[0128] 2. DNA bisulfite conversion
[0129] The DNA bisulfite conversion kit was purchased from Zymo, and was carried out according to the instructions of the kit.
[0130] 3. Multiplex PCR amplification
[0131] Using primer pairs for 22 methylated regions (SEQ ID NO.1-22), multiplex PCR was carried out in one reaction well (see Table 2 for the primer sequence, and the sequence in Table 2-2 was used in this embodiment) to amplify The target sequence containing the target region is produced, and the prod...
Embodiment 3
[0154] Parallel detection of co-methylation of any 1-3 methylated regions among 22 methylated regions
[0155] When the co-methylation of the target methylated region is detected in parallel as any 1-3 of the 22 methylated regions, using the combination scheme in Table 4 in Example 2, the following detection methods can be used. The specific detection process is as follows:
[0156] 1. DNA extraction
[0157] The extraction kit was purchased from QIAGEN, and was carried out according to the instructions of the kit.
[0158] 2. DNA bisulfite conversion
[0159] The DNA bisulfite conversion kit was purchased from Zymo, and was carried out according to the instructions of the kit.
[0160] 3. Fluorescent quantitative PCR assay
[0161] Select the primers and probes of 1-3 methylated regions and the primers and probes of the internal reference, and measure them in one reaction well (see Table 2 for the sequences of primers and probes, and see Example 2 for the combination sche...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com