DNA methylated biomarker related to prognosis of renal clear cell carcinoma
A technology of renal clear cell carcinoma and biomarkers, which can be applied in the field of biomedicine to solve problems such as affecting physiological and pathological activities and affecting gene expression.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] Example 1. Analysis of whole-genome DNA methylation
[0061]DNA methylation basic data collection: 320 clear cell renal cell carcinoma samples were downloaded from the TCGA database, including the following information: clinicopathological factors, such as age and stage at diagnosis, lymph node status, and race and ethnicity, etc.:
[0062] Microarray data: The gene expression dataset (GSE66272) was analyzed using the Gene Expression Profile (GEO, https: / / www.ncbi.nlm.nih.gov / geo / ) of the National Center for Biotechnology (NCBI). Among them, GSE66272 includes 27 pairs of renal clear cell carcinoma tissues and their adjacent normal tissues (platform: GPL96 Affymetrix Human GenomeU133A Array).
Embodiment 2
[0063] Example 2. Analysis of the relationship between differentially expressed CpG sites and differentially expressed genes between ccRCC and adjacent normal tissues
[0064] Methylation data processing: use the TCGA level-3 database to analyze the β value of the relevant CpG site and the location and chromosome of the CpG site;
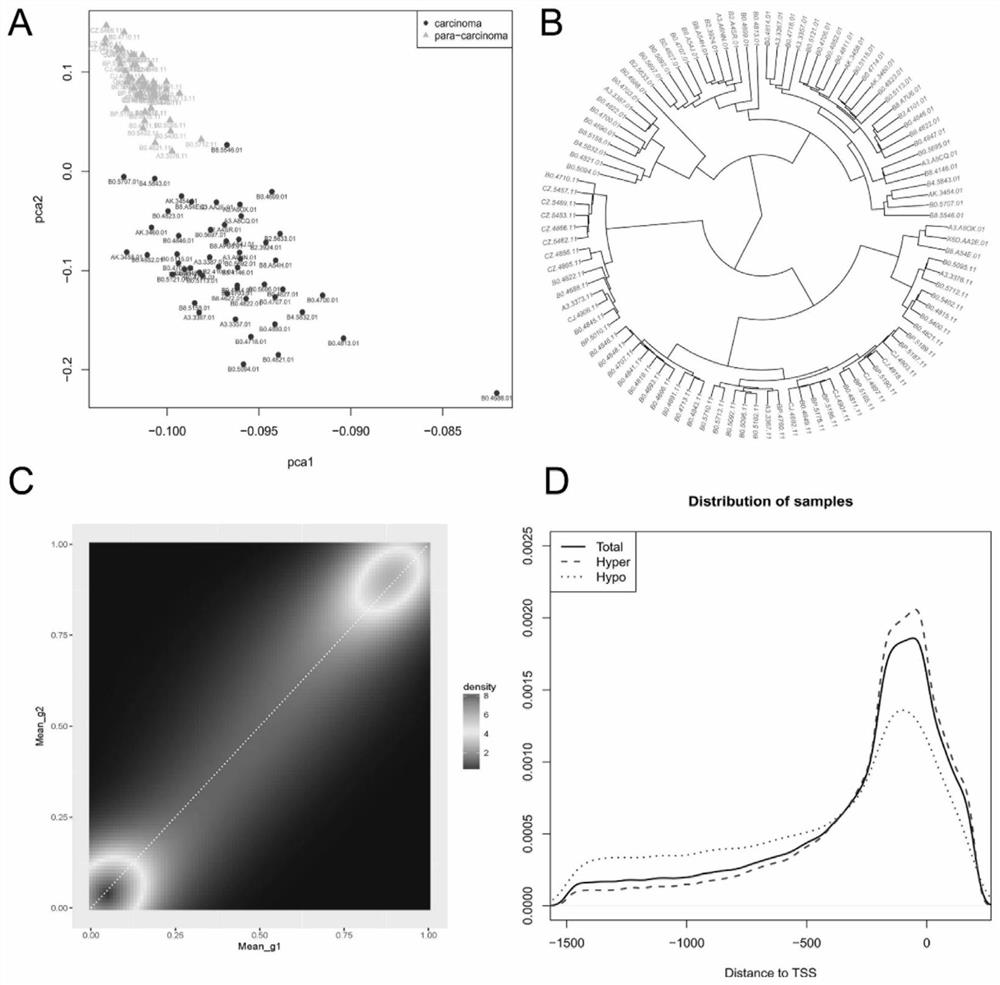
[0065] The β value is calculated by (M / M+U), and its numerical range is from 0 to 1, wherein, M represents methylation, U represents unmethylation, and the present invention removes the CpG probe on chromosome X and Y In order to eliminate gender differences, the methylation level of 102 samples was detected at the CpG site and gene level, and the 102 samples included 51 primary tumor tissues and 51 normal tissues;
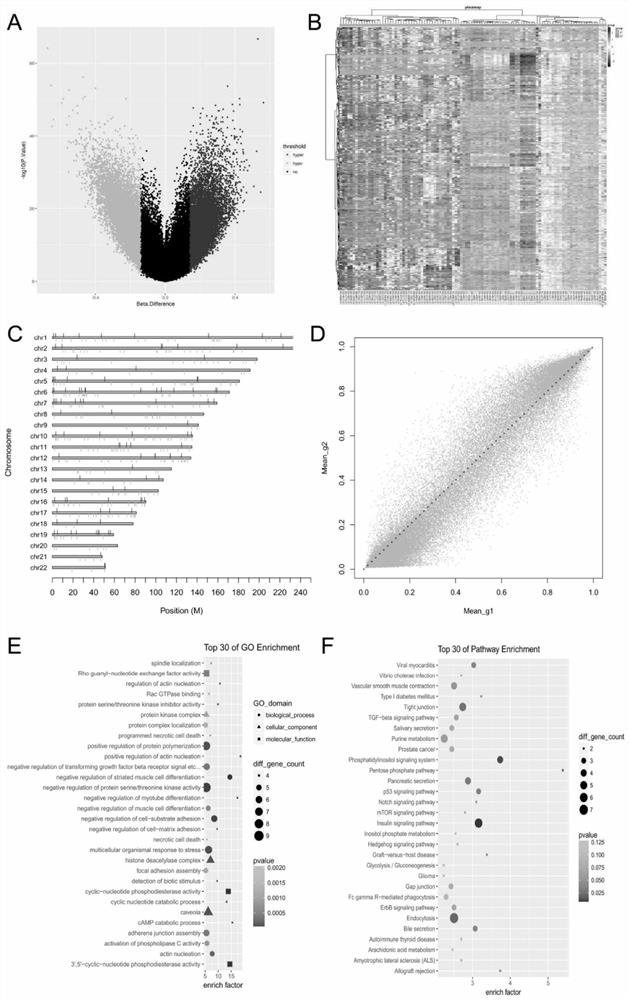
[0066] Differential methylation site analysis: All probes including CpG single nucleotides within 10 bp in length are removed, when the β value between the primary tumor and normal tissue samples is greater than or equal to 0.14, and th...
Embodiment 3
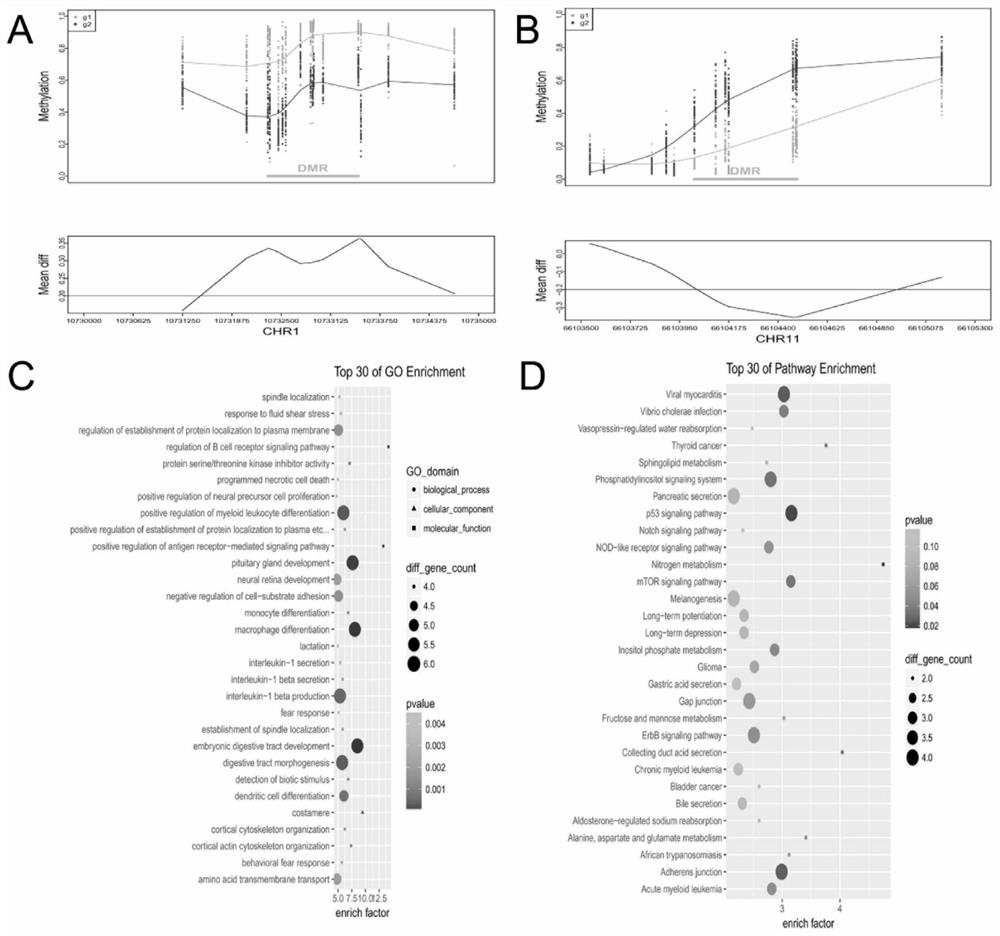
[0072] Example 3. Screen out CpG sites as biomarkers and analyze their relationship with survival
[0073] LASSO regression analysis: For high-dimensional data, LASSO is used to optimize the selection of genes to prevent overfitting and obtain higher diagnostic and prognostic values. LASSO makes the coefficient value close to 0 by eliminating redundant variables. LASSO regression analysis was performed using R software version 3.0.3 and the "glmnet" package;
[0074] Data analysis: use R software to analyze the data, the significant feature is that the two-sided P value is <0.05, and the data is represented by the mean ± standard deviation or the median. In the regression analysis, the P value is calculated by the likelihood ratio test , using T-test to analyze continuous data, chi-square test to compare proportions;
[0075] Use non-zero coefficients in LASSO logistic regression to identify candidate CpG sites;
[0076] Use the single factor Cox proportional hazards regress...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com