Improved gene editing system
A gene editing and genome technology, applied in the field of genetic engineering, can solve problems such as inaccurate mutation types, reduced efficiency, and unpredictability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0139] Example 1. Construction of a gene editing system (ACD) for precise short fragment deletion
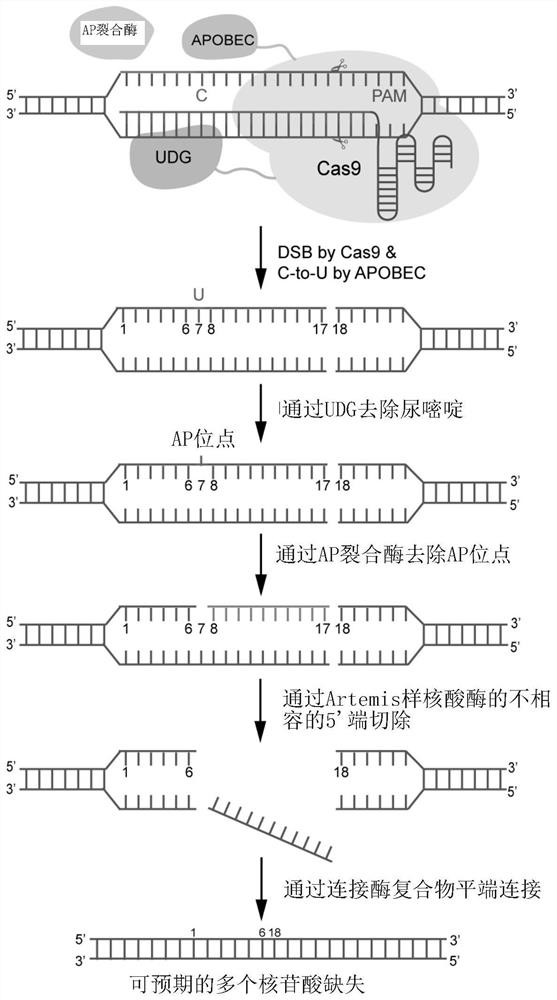
[0140] A single base editing system has been established in 2016 (Komor et al., 2016; Ma et al., 2016; Nishida et al., 2016). The system uses nCas9 (D10A) to guide cytosine deaminase to act on the non-complementary strand of the DNA target site, and deaminates cytosine (C) in a specific region to uracil (U), uracil (U) During DNA replication, it will be replaced by thymine (T), thus achieving precise single base substitution of C-to-T. In the repair process of animal and plant organisms, uracil-DNA glycosylase (Uracil-DNA Glycocasylase, UDG) will preferentially recognize the U base and remove the N-glycosidic bond of the base to form an apurinic or apyrimidinic site (apurinic or apyrimidinic site, AP site), and then under the action of AP lyase (AP lyase), the U base is repaired into the original C base by base excision repair. Therefore, Uracil-DNA Glycocasylase inhibitor (UG...
Embodiment 2
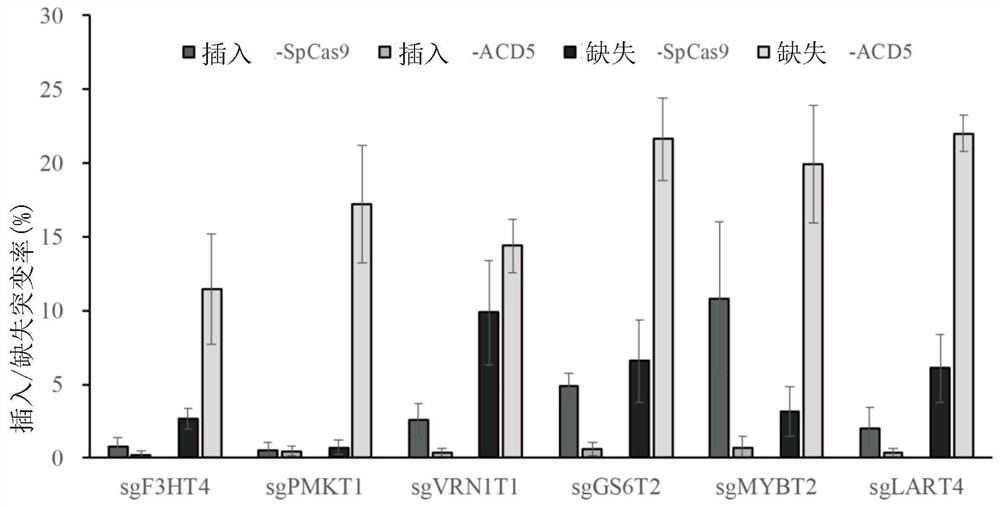
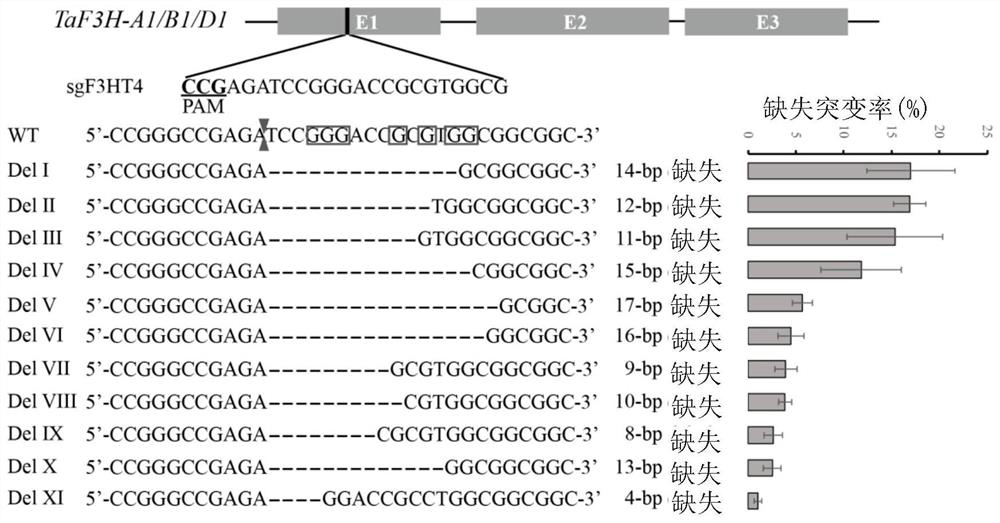
[0143] Example 2, the analysis of the missing type produced by the ACD system
[0144] Sequence analysis of the Deletion mutations generated by the ACD system on different target sites ( Figure 3-8 ), except for a few types, most of the mutation types are in line with expectations, all of which are APOBEC3A action bases (NGG (PAM) corresponds to C bases; CCN (PAM) corresponds to G bases) to the Cas9 cleavage site Deletion between points. However, due to the asymmetry of Cas9 in cutting the double strand, Cas9 will cut between the 3-4 or 4-5 near the PAM end. In addition, the bases that APOBEC3A acts on on the non-target strand will use the target strand as a template to form 1-2 bases that pair with the complementary strand during the repair process. Therefore, 1-2 bases that are compatible with the target strand may also be introduced bases paired with complementary strands.
[0145] The efficiency of the ACD system to generate Insertion is very low, but the efficiency of...
Embodiment 3
[0146] Embodiment 3, construct AFID (APOBEC-Cas9 Fusion-Induced Deletion) system
[0147] The present invention selects human APOBEC3A with high deamination activity and wide deamination window to construct the AFID-3 system, and screens a higher deamination activity and narrow window APOBEC3Bctd to replace APOBEC3A to construct the eAFID-3 system ( Figure 9 and Figure 10 ). A comparative analysis of the deletion efficiencies of Cas9, AFID-3 and eAFID-3 on the endogenous gene targets of rice and wheat showed that the efficiency of deletion mutations was significantly increased compared with Cas9, AFID-3 and eAFID-3, and its The average deletion mutation rate is 2.2 times and 2.6 times that of Cas9, which fully demonstrates the high efficiency of the AFID system.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com