ShRNA sequence for specifically inhibiting GOS2 gene expression and application thereof
A gene expression and specific technology, applied in the field of shRNA sequence that specifically inhibits GOS2 gene expression, can solve the problem of short time to function
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] First, based on the GOS2 gene (the accession number in GENABANK is NM_015714.4), four primer pairs corresponding to shRNA were designed, which were recorded as shRNA primers. The specific sequences of the four pairs of shRNA primers are shown in Table 1. The 4 pairs of shRNA primers were annealed to form 4 short double-stranded oligonucleotide fragments to obtain shRNAs, which were named shGOS2-1, shGOS2-2, shGOS2-3, and shGOS2-4, respectively. .
[0040] shGOS2-1: 5'-GGAGCGACAGGCTCTCCAGAA-3' (SEQ ID NO: 1);
[0041] shGOS2-2: 5'-GGCCCTGTCCAACCGGCAGCA-3' (SEQ ID NO: 2);
[0042] shGOS2-3: 5'-GCCGCCAGACGTCTGCGGGAC-3' (SEQ ID NO: 3);
[0043] shGOS2-4: 5'-GCGACAGGCTCTCCAGAAGCA-3 (SEQ ID NO: 4).
[0044] Table 1. Primer pairs corresponding to shRNA sequences
[0045]
[0046] The shGOS2-1, shGOS2-2, shGOS2-3, and shGOS2-4 obtained above were connected to the double-enzyme-cut lentiviral vector Plko.1-TRC (Open Biosystem Company), respectively, and the restriction si...
Embodiment 2
[0048] Example 2: Preparation of cell lentivirus and transfection of cells with lentivirus
[0049] Preparation of Mixture A:
[0050] Solution A of the experimental group: 0.75 µg helper plasmid pLP1 + 0.35 µg helper plasmid pLP2 + 0.49 µg helper plasmid pL PSVG + 0.61 µg target plasmid Plko.1-TRC-shRNA-shGOS2 obtained in Example 1 OpenBiosystem Company, Sangon Bioengineering (Shanghai) Co., Ltd.).
[0051] Solution A of the control group: 0.75 µg helper plasmid pLP1 + 0.35 µg helper plasmid pLP2 + 0.49 µg helper plasmid pL PSVG + 0.61 µg vector plasmid Plko.1-TRC (Open Biosystem).
[0052] Add solution A to 0.125 μL of serum-free low-glucose DMEM medium (purchased from Thermo Fisher), mix gently, and incubate at room temperature for 5 min.
[0053] Preparation of mixture B: Take two 1.5mL EP tubes and add 125 μL of serum-free low-glucose DMEM medium, and then add 9 μL of EZ-Trans cell transfection reagent (purchased from Shanghai Liji Biotechnology Co., Ltd.) Mix lightly ...
Embodiment 3
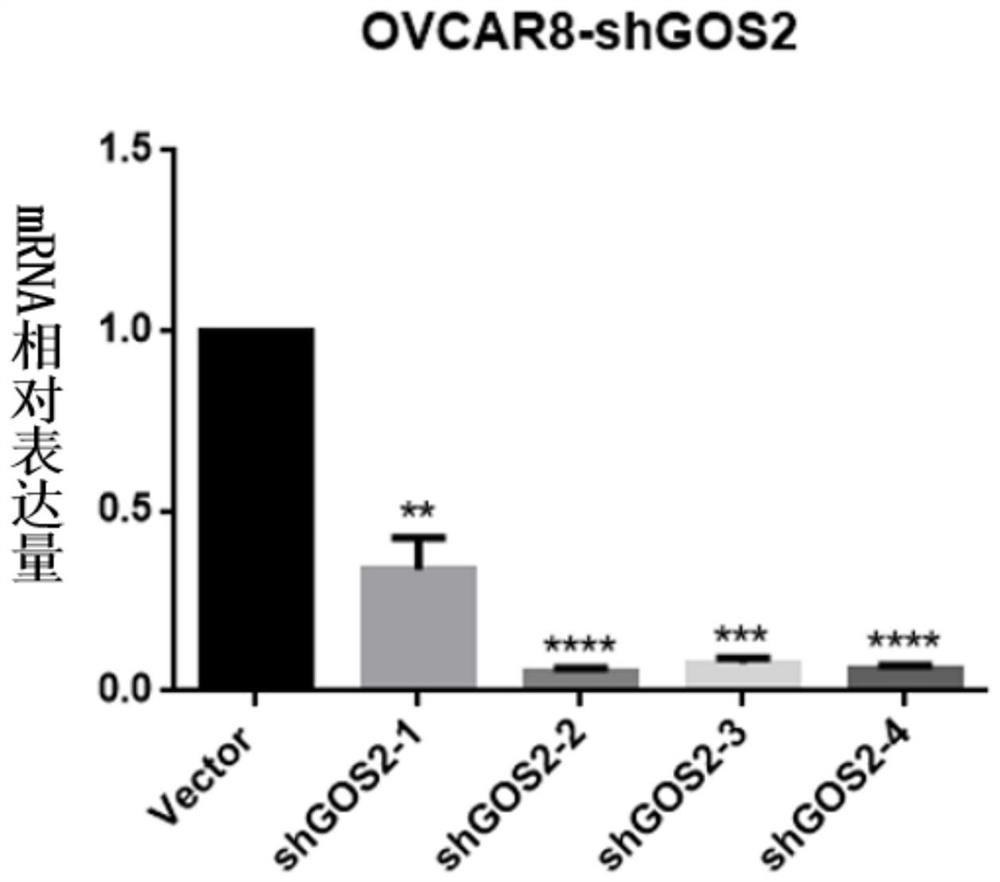
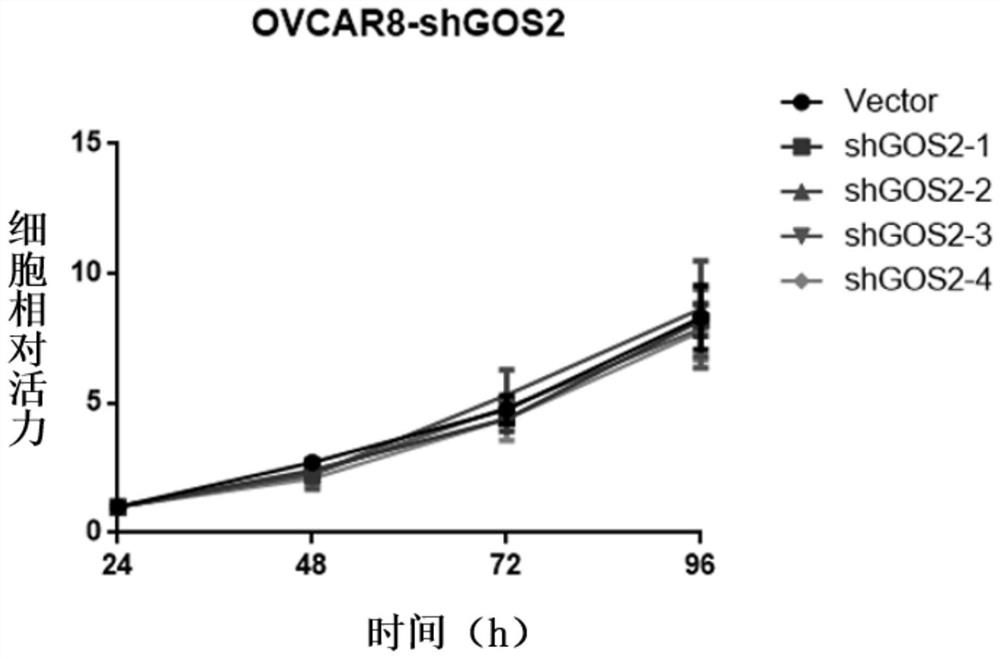
[0059] Embodiment 3: Detection of the expression level of GOS2 gene in OVCAR8 cells
[0060] Collect the surviving OVCAR8 cells in the experimental group and the control group after puromycin (puromycin; PM) screening in Example 2, use the TRIzol method to extract RNA, and then operate according to the instructions of the real-time fluorescence quantitative kit (purchased from Novizyme) to detect The mRNA expression of target GOS2 in different groups of cells.
[0061] According to the experimental results, the Ct values of the experimental group and the control group (the number of cycles experienced when the fluorescent signal in each reaction tube reaches the set threshold value) were obtained using 2 -△△Ct Values were used to calculate the silencing efficiency of shGOS2. Among them, △△Ct=△Ct of the experimental group-△Ct of the control group, △Ct=(Ct of the experimental group-Ct value of the internal reference of the experimental group) / (Ct of the control group-Ct of ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com