Bacterial genome multi-editing method based on double-stranded DNA recombinant engineering and application thereof
A DNA recombination and genome technology, applied in genetic engineering, recombinant DNA technology, DNA/RNA fragments, etc., can solve the problems of establishment of multiple genome editing methods, low efficiency, low editing efficiency, etc., to achieve low cost of substrate preparation, enhanced Strain optimization, the effect of unlimited length of inserted gene
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0069] Embodiment 1: A dsDNA-recombineering assisted Multiplex Genome Editing (dReaMGE) method applicable to a variety of bacteria provided by the present invention, the specific implementation steps are as follows:
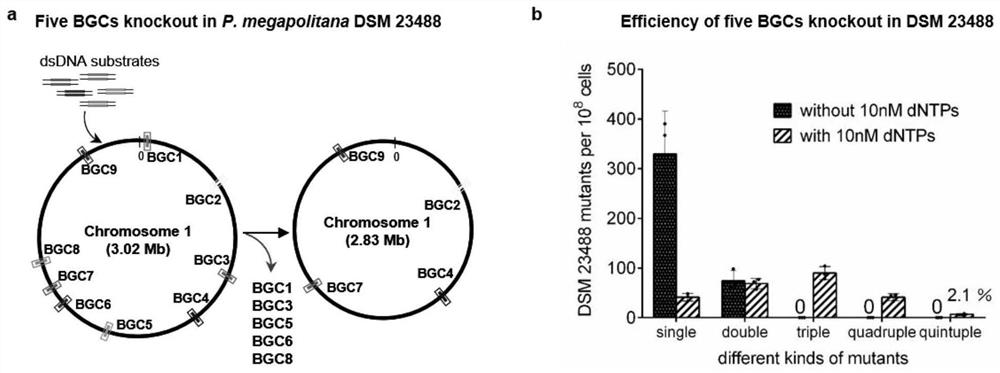
[0070] (1) Select Red / ET type recombinase that can effectively mediate recombineering for different hosts, suitable inducible promoters, strong promoters, resistance markers and backbone plasmids; wherein, the hosts include Escherichia coli E.coliGB2005, primary Five different bacteria including Polyangium brachysporum DSM 7029, Burkholderia Paraburkholderia.megapolitana DSM 23488, Pseudomonas.putida KT2440 and Pseudomonas.syringae DC3000.
[0071] (2) Use the inducible promoter screened in (1) to control the recombinase and construct it on a suitable backbone plasmid, and convert the constructed gene into The recombinant enzyme expression plasmid is introduced into the host cell.
[0072] (3) A dsDNA substrate with short homology arms (100 bp) capable of anchor...
Embodiment 2
[0080] Example 2: dsDNA-mediated simultaneous editing of dual regions of the E.coli genome
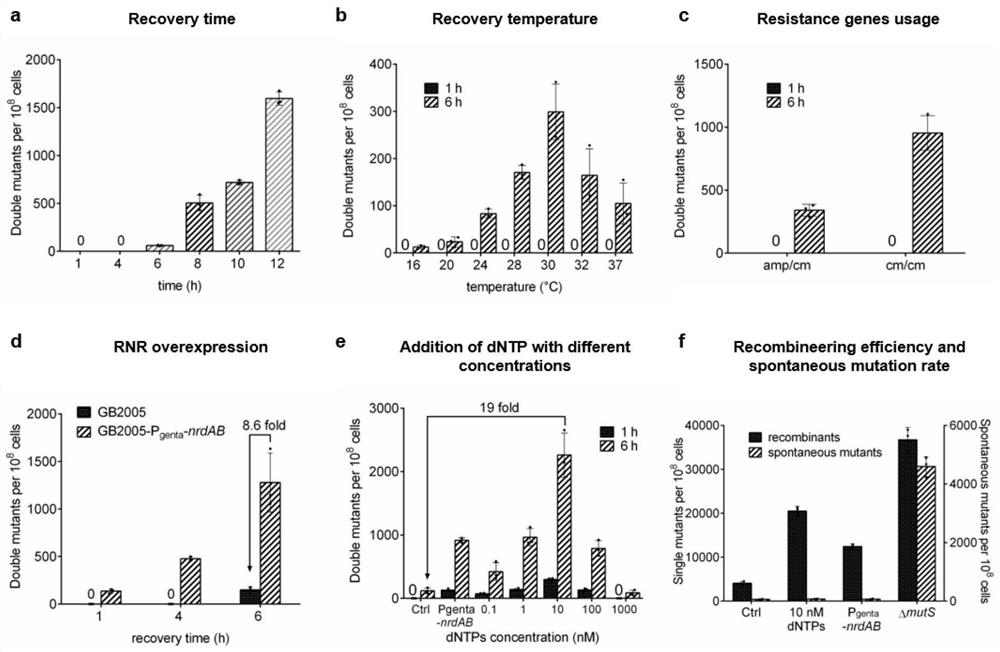
[0081] In Escherichia coli GB2005, it was verified that the dsDNA recombineering of the present invention mediates the simultaneous editing of two genome regions at different recovery times and different recovery temperatures.
[0082] Primer sequence: the lowercase part is used to mediate sufficient homology arms, and the uppercase part is the primer for amplifying the resistance gene.
[0083] Pgenta-nrdAB-DH10B-3: gcagtccttctgccgCccaatccagaacgcgatgGattttgtcgagattgatgcg ctctgtgctaccgtcgcgctttgtcaccagcagattctgattcatATGTATATCTCTTTAGGTG;
[0084] P genta -nrdAB-DH10B-5: tcgcttatatattgaccacaactgatacatcagattatgtgatgactcgtgcttagatca atttttgcaatcattagcaaaaagattaataagccatctaAGGCACGAACCCAGTTGACA;
[0085] GB2005-regionA-delet-3: tgcacttcctgccggatatctacgtgccgtgcgaccagtgcaaaggtaaacgc tataaccgtgaaacgctggagattaagtacaaaggcaaaaccatccaTTACGCCCCGCCCTGCCACT;
[0086] GB2005-regionA-delet-5: gctgcca...
Embodiment 3
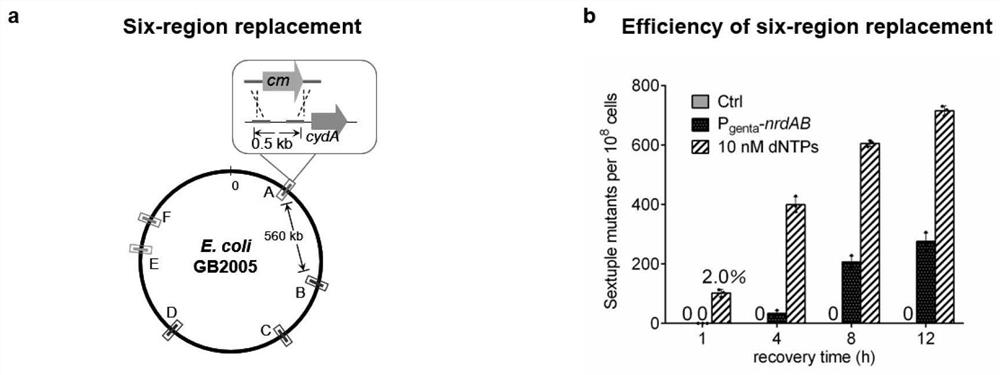
[0095] Embodiment three: Six regions are replaced simultaneously in Escherichia coli GB2005
[0096] This example tests dsDNA recombineering in wild-type E.coli GB2005 and GB2005-P genta -In nrdAB, the ability to mediate the simultaneous replacement of six regions of the genome (0.5kb) under the condition of adding / not adding dNTPs, such as figure 2 a.
[0097] Primer sequence:
[0098] GB2005-regionC-delet-3: attgaagcagaagcctgcgatgtcggtttccgcgaggtgcggattgaaaatggt ctgctgctgctgaacggcaagccgttgctgattcgaggcgttaaccTTACGCCCCGCCCTGCCACT;
[0099] GB2005-regionC-delet-5: gccagctggcagttcaggccaatccgcgccggatgcggtgtatcgctcgccactt caacatcaacggtaatcgccatttgaccactaccatcaatccggtCGTTGATCGGCACGTAAGAG;
[0100] GB2005-regionD-delet-3: cgcctaatacatctacactttctatttattgacaagtgatacgttgcaaaaggagcaacacccccacagactcgatgactgcgcagtcatacagtgaaattTTACGCCCCGCCCTGCCACT;
[0101] GB2005-regionD-delet-5: taggaatttcggacgcgggttcaactcccgccagctccaccaaaattctccatcgg tgattaccagagtcatccgatgaagtcctaagagcccgcacggcCGT...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com