Method for differentiating natural killer cells from pluripotent stem cells based on single cell sequencing rational design and application of CSF1R inhibitor
A technology of natural killer cells and pluripotent stem cells, applied in the field of high-resolution cell atlas, can solve the problems of long differentiation time, blind optimization of differentiation methods, and unstable system, and achieve the effect of promoting differentiation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0053] The method for preparing natural killer cells provided by the present invention, simple and convenient, can be stably, and quickly obtain high purity natural kill cells.
[0054] In some preferred embodiments, the CSF1R inhibitor includes, but is not limited to, BLZ 945 and / or PLX3397, for example, LINIFANIB (ABT-869), OSI-930, GW2580, PLX5622, and the like. In the present invention, it is possible to suppress the expression of CSF1R in the cells and does not affect the substance of cell survival.
[0055] It should be noted that "BLZ945 and / or PLX3397" means that the CSF1R inhibitor may include only BLZ945, or may include only PLX3397, and may be BLZ945 and PLX3397.
[0056] In some preferred embodiments, the step of obtaining multi-capable cell induction culture to obtain a pyrogenic bobbin comprises performing a plurality of stem cells in the first medium;
[0057] The first medium includes: base medium, Y27632 8 to 12 μm, BMP416 to 24 ng / ml, VEGF 16 to 24 ng / ml,...
Embodiment 1
[0075] Preheat (15-25 ° C) is sufficient amounts of MTESR1.
[0076] IPS cells were digest into single cells with Tryple.
[0077] 1. Use 1ml without CA 2+ And MG 2+ PBS wash original hole.
[0078] 2. Insert PBS and add 200 μL of tryple to digest for 5 minutes.
[0079] 3. Centrifuge 200 × g after digestion for 5 minutes.
[0080] 4. Revenue with the following medium:
[0081] Basic medium: STEMDIFF TMApel TM 2 contained Y27632 (10 μM), BMP4 (20 ng / mL), VEGF (20 ng / mL) and SCF (40 ng / mL).
[0082] 5. Adjust the cell density to 80,000 tells / ml, add 100 μl / pore cell suspension in low adsorbed 96-well plates.
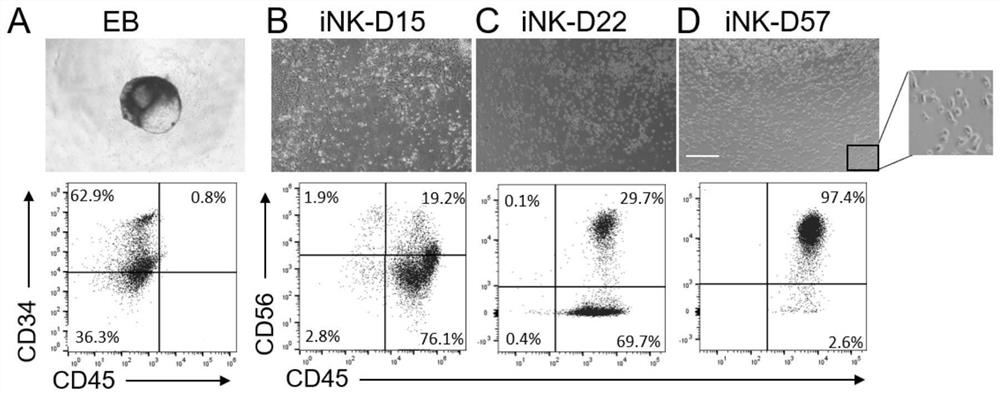
[0083] 6.300 × g Centrifuge for 5 minutes in 37 ° C, 5% CO 2 The cell culture box was cultured for 6 days, and EB (ambugula), on day 7, 20-40 EBs were placed in a 6-well plate having a matrix glue (1 mg / ml or 0.1% degass), with the following Culture culture medium:
[0084] Basic medium: STEMSPAN TM -XF, containing SCF (20 ng / ml), IL-7 (20 ng / mL), IL-15 (10 ng...
Embodiment 2
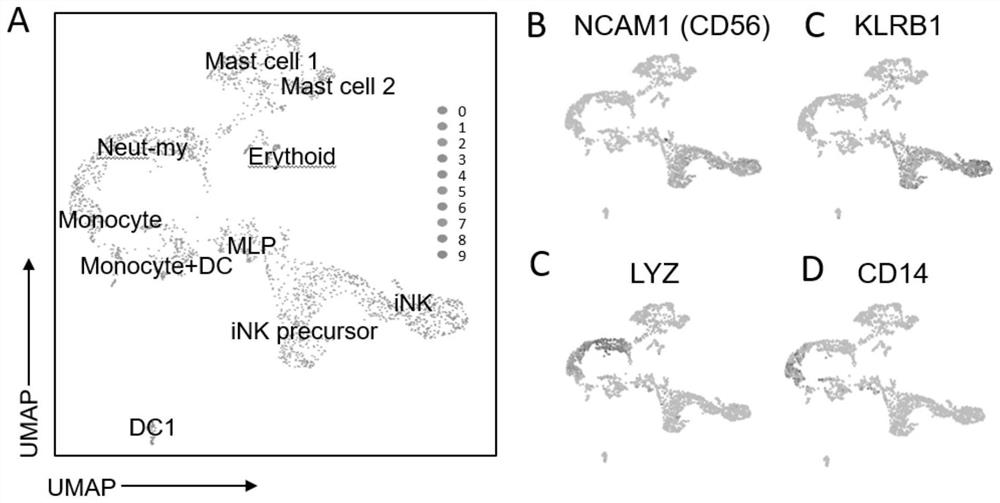
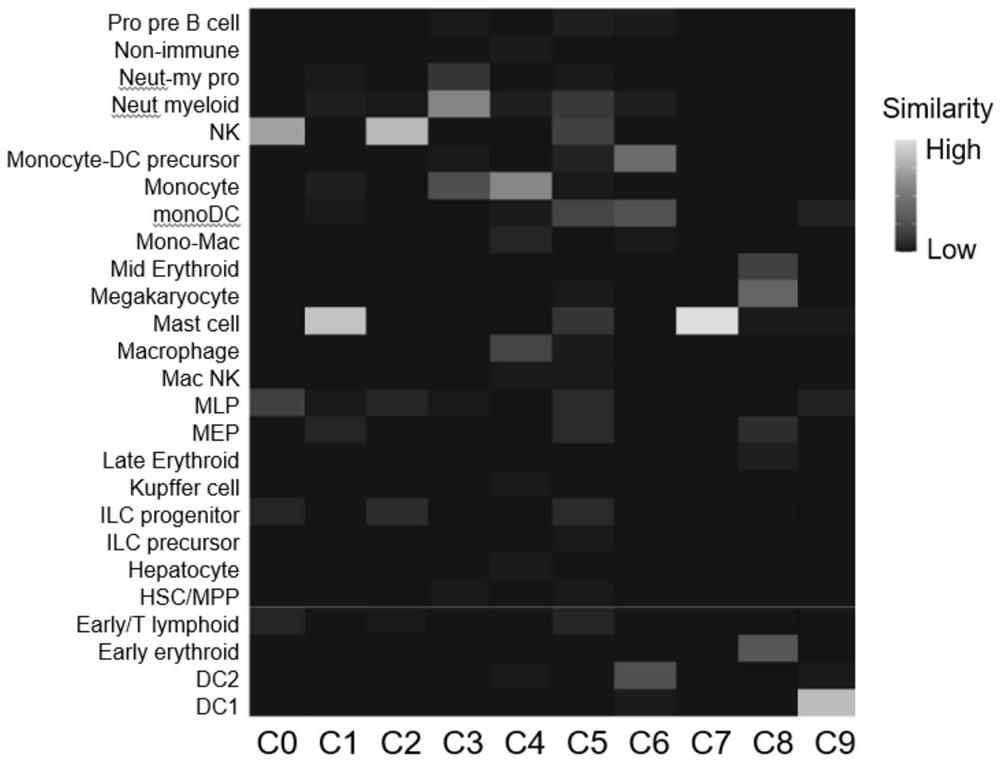
[0088] The cells were collected in Example 1 to last 30th day, and the CSF1R inhibitor BLZ9450.5 μm and PLX3397 1 μm were treated, respectively, and the control group (not treated, normal cultured) was provided, and the CD45 was collected and detected after 8 days. + CD56 + The ratio of NK cells, Figure 4 Indicated (shown) Figure 4 The expression of A is NK differentiation to the 24th day of the CSF1R in the UMAP map, Figure 4 B is CSF1R changes in the amount of mRNA expression during macrophages in vitro, and in vitro. Figure 4 C is the results of control group flow cytometry analysis, Figure 4 D is the result of the analysis of the BLZ945 treatment of group flow cytometry. Figure 4 The E is the results of PLX3397 treatment of group flow cytometry analysis.
[0089] Such as Figure 4 The A, CSF1R is highly expressed in the in vitro differentiation system in myeloid cells (mononuclear cells, dendritic cells, etc.); almost no expression in strand cells (NK cell precursors, NK cells,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com