Python data fusion-based Tibetan medicine meconopsis integrifolia producing area discrimination method
A technology of Meconopsis entire margin and data fusion, applied in character and pattern recognition, measuring devices, material analysis through optical means, etc., can solve the limitations of inability to fully reflect the complex chemical composition of Tibetan medicinal materials, origin traceability and quality evaluation issues such as sex, to achieve the effect of improving the accuracy of origin discrimination and the recognition effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] In an exemplary embodiment, there is provided a method for discriminating the origin of Tibetan medicine Meconopsis entire margin based on Python data fusion, such as figure 1 As shown, the method includes:
[0055] Samples of Meconopsis whole-leaf from different origins were collected;
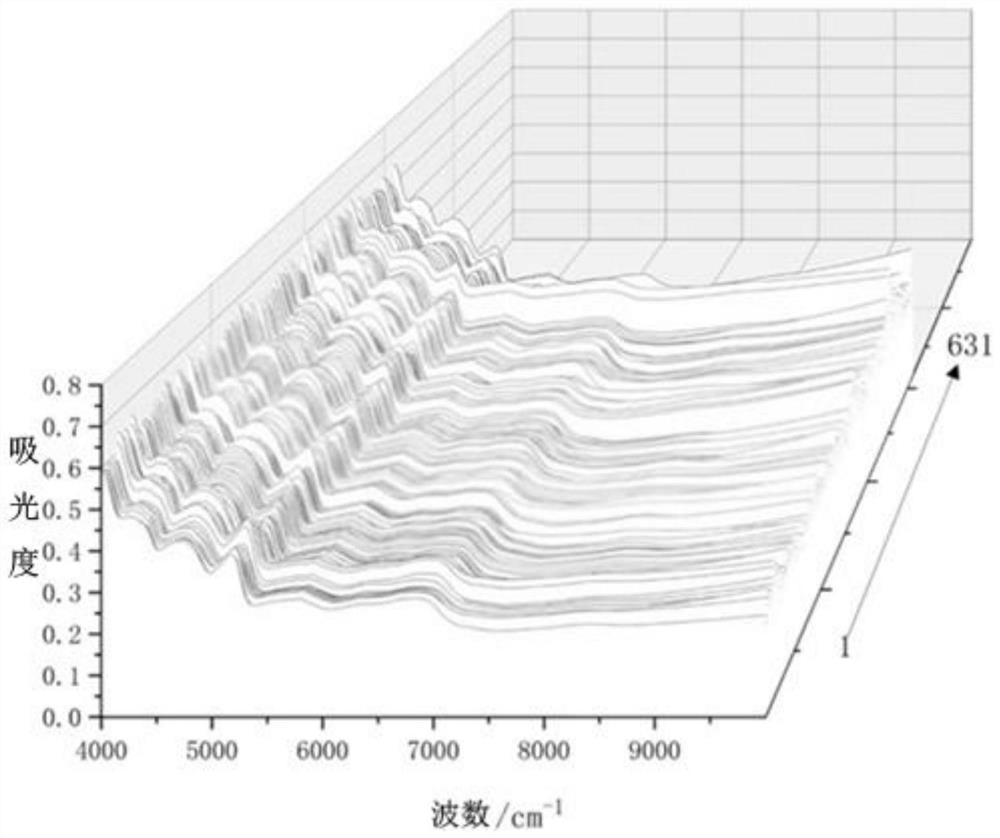
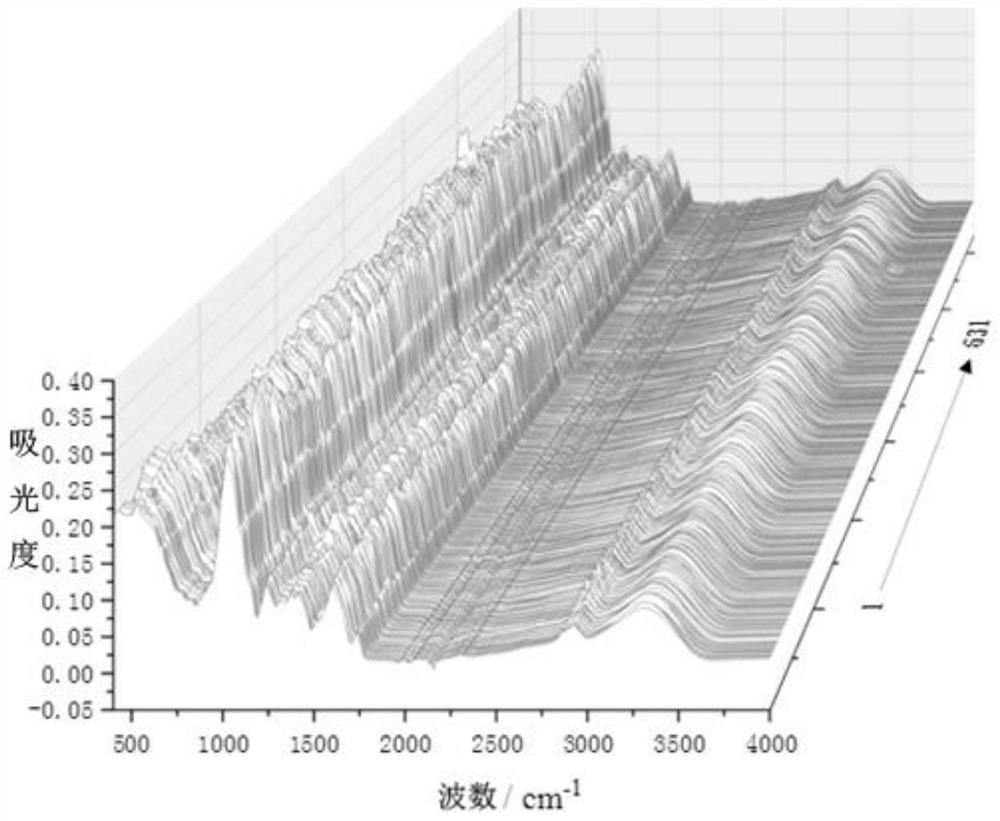
[0056] Collecting NIR spectra of the samples of Meconopsis entire from different origins to obtain NIR spectral data, and collecting ATR spectra on the samples of Meconopsis entire from different origins to obtain ATR spectral data;
[0057] Fusing the NIR spectral data and the ATR spectral data to obtain primary fusion data; using multiple classification methods to model the primary fusion data respectively, and comparing the classification effects of different models to obtain the classification method with the best classification effect;
[0058]Use Python software to extract the eigenvalues of each spectral data in the primary fusion data, calculate the contribution of each eige...
Embodiment 2
[0064] Based on Example 1, a method for discriminating the origin of Tibetan medicine Meconopsis whole margina based on Python data fusion is provided, and the method also includes:
[0065] Select several classification methods with higher classification effects to model the intermediate fusion data respectively, and perform high-level fusion of the output results of multiple models, and the high-level fusion includes:
[0066] Weights are assigned to the output results of various models, a new decision-making method is constructed, and the physical and chemical properties of the sample molecules are analyzed according to the decision-making method. Among them, the output results of multiple models are voted, and each model obtains a voting prediction result, and then fuses the voting results to form a model to complete advanced fusion. This model can be called an advanced discriminant model. Using this advanced discriminant The model discriminates the origin of Meconopsis en...
Embodiment 3
[0069] Based on Example 1, a method for discriminating the origin of Tibetan medicine Meconopsis entire margin based on Python data fusion is provided, and the collection of samples of Meconopsis entire margin from different origins includes:
[0070] A total of 631 samples of Meconopsis whole leaf were collected from 14 different producing areas in Qinghai Province. Specifically, during the flowering period of Meconopsis entire margin, a total of 631 whole plant samples were collected from 14 different producing areas from south to north in Qinghai Province. The samples were first identified as Meconopsis entire margin, The samples were taken back to the laboratory to be washed, dried, crushed, passed through a 100-mesh sieve, and put into a desiccator for analysis.
[0071] Experimental equipment includes: iS50 Fourier Transform Infrared Spectrometer (Thermo Fisher, USA), equipped with near-infrared and ATR accessories, a sample cup with a diameter of 1.20 cm, and a desiccat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com