Construction method of pseudomonas denitrificans lacking malonic semialdehyde bypass metabolic pathway
A technology of Pseudomonas denitrification and malonate semialdehyde, applied in the biological field, can solve the problems of low yield and achieve the effect of increasing yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
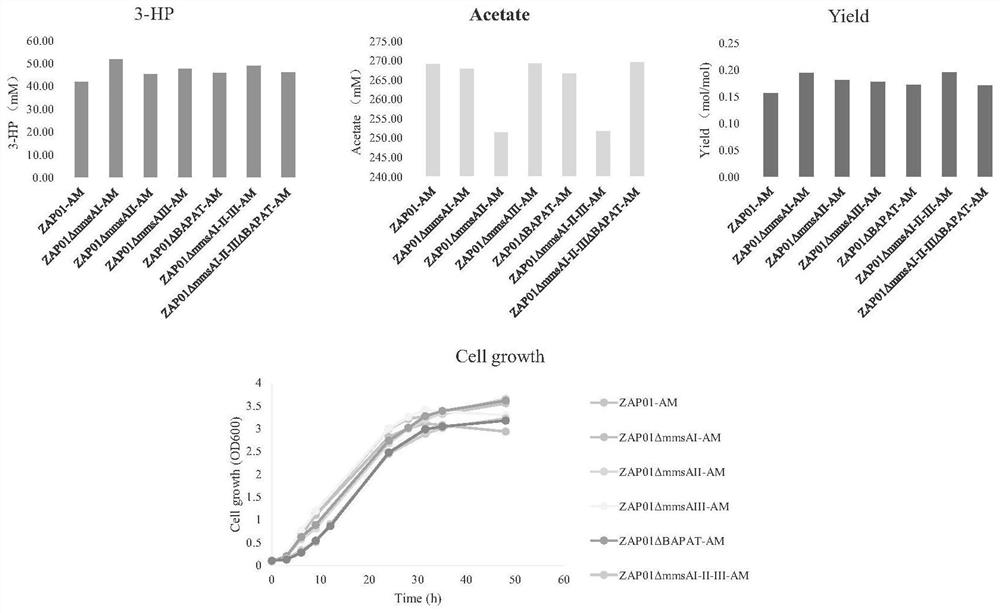
[0026] 1. Knockout of malonate semialdehyde bypass metabolic pathway
[0027] The starting strain of the present invention is Pseudomonas denitrificans ZAP01 constructed in the previous stage. The mmsAI, mmsAII, mmsAIII and BAPAT genes were knocked out separately and in combination in the genomic DNA of the strain to obtain mutant strains ZAP01ΔmmsAI, ZAP01ΔmmsAII, ZAP01ΔmmsAIII, ZAP01ΔBAPAT, ZAP01ΔmmsAIΔmmsAIIΔmmsAIII and ZAP01ΔmmsAIΔmmsAIIΔmmsAIIIΔBAPAT.
[0028] In the present invention, the above-mentioned genes are knocked out by using the technique of a traceless gene knockout. The specific operation is as follows (taking the knockout of the gene mmsAI as an example):
[0029] For the knockout of the gene mmsAI, at first the DNA fragment mmsAI US (SEQ ID No.2) of about 700bp upstream sequence of the mmsAI gene (SEQ ID No.1) was cloned using the genomic DNA of Pseudomonas denitrophila ATCC13867 as a template, and mmsAI The DNA fragment mmsI DS (SEQ ID No.3) of about 700...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com