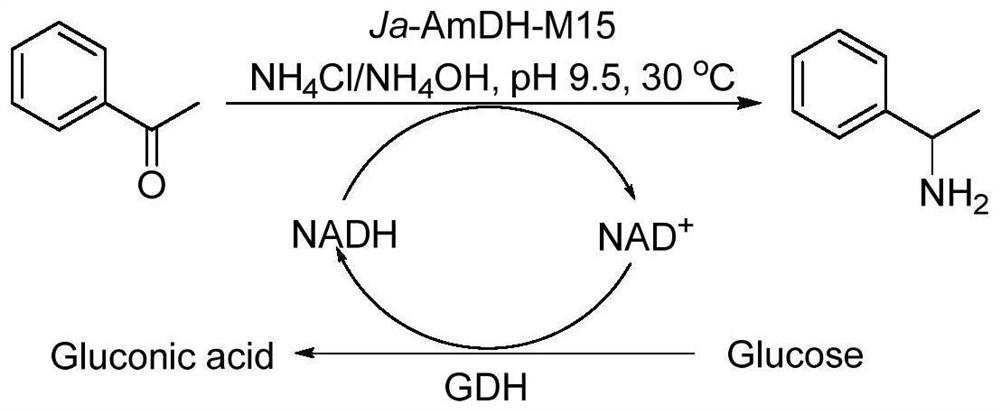
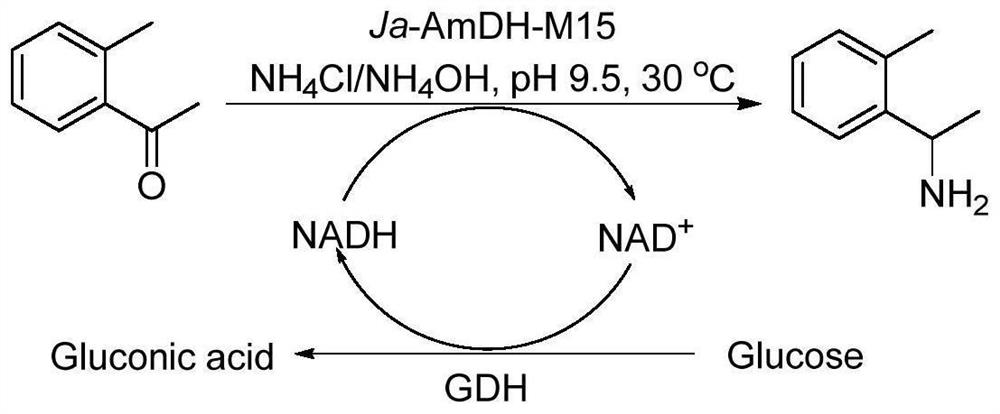
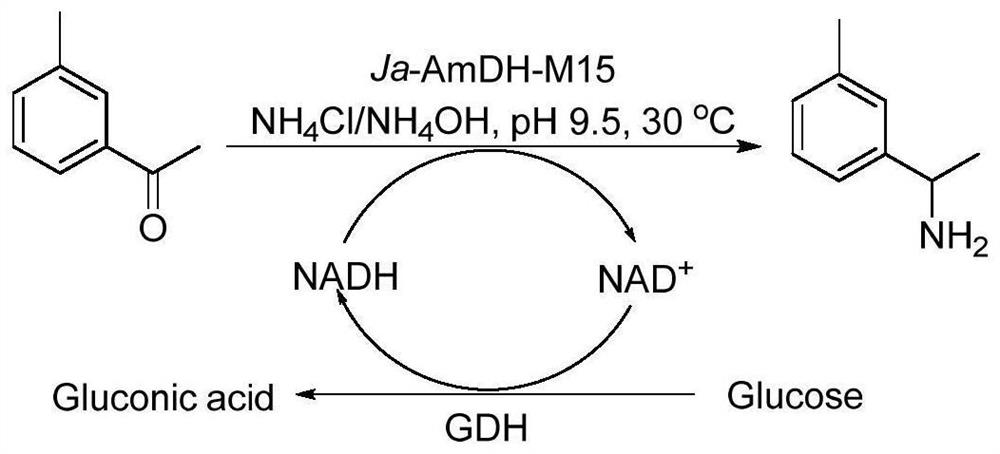
Amine dehydrogenase as well as coding nucleic acid and application thereof
A technology of amine dehydrogenase and amino acid dehydrogenase, applied in amine dehydrogenase, the application field of amine dehydrogenase, can solve the problems of narrow substrate spectrum, few kinds of AmDH, limited industrial application, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0106] Example 1, Screening and Determination of Mutation Sites
[0107] The natural amino acid dehydrogenase protein derived from Jeotgalicoccus aerolatus (as shown in SEQ ID NO: 2, its coding gene (AmDH gene) as shown in SEQ ID NO: 1) was subjected to sequence analysis, mutation and functional verification, specifically selected 12 amino acid sites, and 4 important amino acid sites were screened out from the 12 amino acid sites through further research and analysis. The four amino acid sites are mutated in different forms to obtain mutant proteins.
[0108] The four amino acid sites and their mutant forms are shown in Table 1.
[0109] Table 1: 15 mutant forms
[0110] mutant number mutant form a1 K67S a2 K67Q a3 K67M a4 N260L a5 N260C a6 N260F a7 N260I a8 K67S\N260L a9 K67S\N260C a10 K67S\N260F a11 K67S\N260I a12 K67S\N260L\E113V a13 K67S\N260L\E113V\V291A a14 K67S\N260L\E113V\...
Embodiment 2
[0119] Embodiment 2, the preparation of recombinant bacteria
[0120] 1. Construction of wild-type recombinant expression vector
[0121] The nucleotide sequence shown in SEQ ID NO: 1 was inserted between the EcoRI and XhoI restriction sites of the pET28a vector to obtain the recombinant expression vector pET28a-AmDH, which was verified to be correct by sequencing.
[0122] 2. Construction of mutant recombinant expression vector
[0123] The nucleotide sequences encoding the mutants a1-a15 (see Table 2) were respectively inserted between the EcoRI and XhoI restriction sites of the pET28a vector to obtain the recombinant expression vector pET28a-1-15, which was verified to be correct by sequencing.
[0124] 3. Preparation of recombinant bacteria
[0125] 1. Transform Escherichia coli BL21(DE3) with the recombinant expression vector pET28a-AmDH obtained in step 1 to obtain wild-type recombinant bacteria.
[0126] 2. Transform the recombinant expression vector pET28a-1-15 prep...
Embodiment 3
[0127] Embodiment 3, the enzyme activity assay of amino acid dehydrogenase mutant protein
[0128] The wild-type recombinant bacteria and mutant recombinant bacteria 1-15 obtained in Example 2 were cultivated, the protein was extracted, and the enzyme activity was detected.
[0129] 1. Inoculate the recombinant bacteria in 10 mL of LB liquid medium containing kanamycin at a final concentration of 50 μg / mL, shake and culture at 37° C. and 180 rpm for 16 hours to obtain seed liquid. Then the seed solution was inoculated into 50 mL of TB medium (inoculum size 1%), containing a final concentration of 50 μg / mL kanamycin in the medium, and cultured with shaking at 37° C. and 180 rpm until the OD of the bacterial solution 600nm 0.6-0.8, add IPTG with a final concentration of 0.5 μM, continue shaking culture at 20°C and 180rpm for 20-24h, and centrifuge at 5000rcf for 15min to collect the bacteria.
[0130] The recombinant bacterial cells were washed and resuspended twice with 50mM 5...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com