SNP (Single Nucleotide Polymorphism) molecular marker linked with peanut bacterial wilt resistant major QTL (Quantitative Trait Loci) and application thereof
A molecular marker, anti bacterial wilt technology, applied in the determination/inspection of microorganisms, recombinant DNA technology, biochemical equipment and methods, etc., to achieve the effect of improving breeding efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
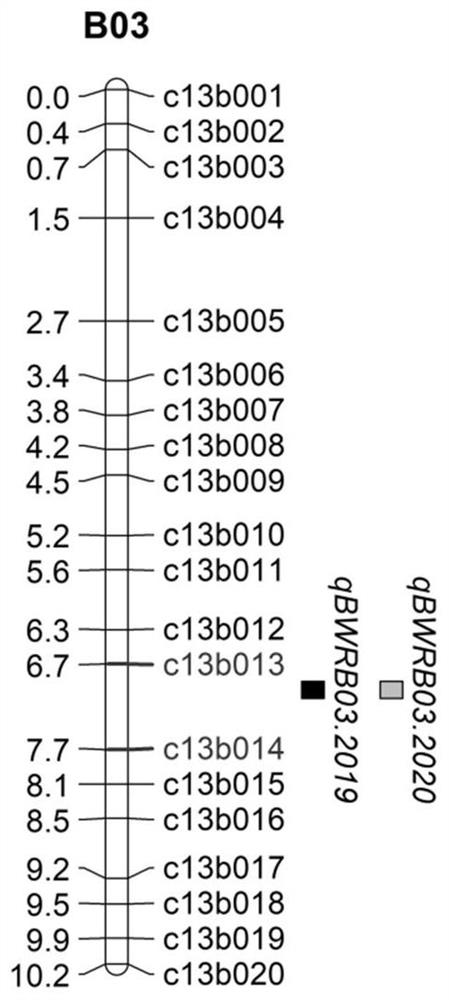
[0036] This example provides the development process of the SNP molecular marker BWRB03-R linked to the main QTL site qBWRB03 for resistance to bacterial wilt in peanut, as follows.
[0037] Test materials: A peanut bacterial wilt-susceptible variety Zhonghua 10 was used as the female parent, and a peanut bacterial wilt-resistant germplasm ICG12625 was used as the male parent to obtain a recombinant inbred line (RIL )group.
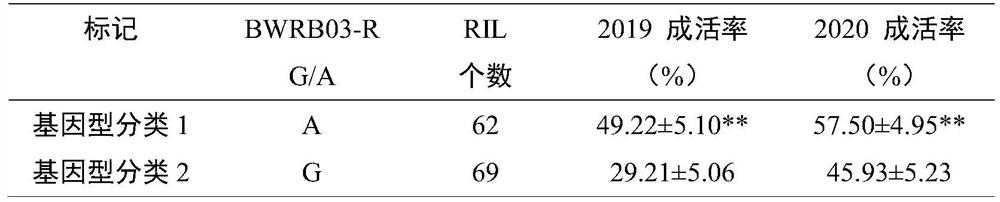
[0038] Phenotype identification: In 2019 and 2020, the bacterial wilt resistance of 140 strains of Zhonghua 10, ICG12625 and RIL populations was identified in the Hongan bacterial wilt disease nursery of the Institute of Oil Crops, Chinese Academy of Agricultural Sciences. A completely randomized block experimental design was adopted, with 3 repetitions. Each time, each material was planted in a row of 20 plants, with a row spacing of 30 cm and a plant spacing of 10 cm. Adopt standard field management practices. After all the seedlings emerged, the tot...
Embodiment 2
[0045] This example provides the 131 RILs of ICG12625 and Zhonghua 10 that obtained survival rate data for both environments in Example 1 by using the SNP molecular marker BWRB03-R linked to the major bacterial wilt resistance locus qBWRB03 to test. Specific steps are as follows.
[0046] Using genomic DNA as a template and the sequences shown in SEQ ID NO.2-3 as primers, the SNP molecular marker BWRB03-R was amplified with the KAPA2G Fast MultiplexMix kit. The PCR conditions were: 95°C pre-denaturation for 3 minutes; 95°C denaturation for 15 s, 55°C renaturation for 30 s, 72°C extension for 30 s, a total of 30 cycles; finally 72°C extension for 5 min, 4°C incubation. After the amplification product was added with a sequencing adapter, the Illumina HiSeq platform was used for Paired-end 150bp (PE150) sequencing, and the sequencing sequence was compared to the reference sequence to detect the base of the SNP site. If the RIL is consistent with the base of the SNP site detecte...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com