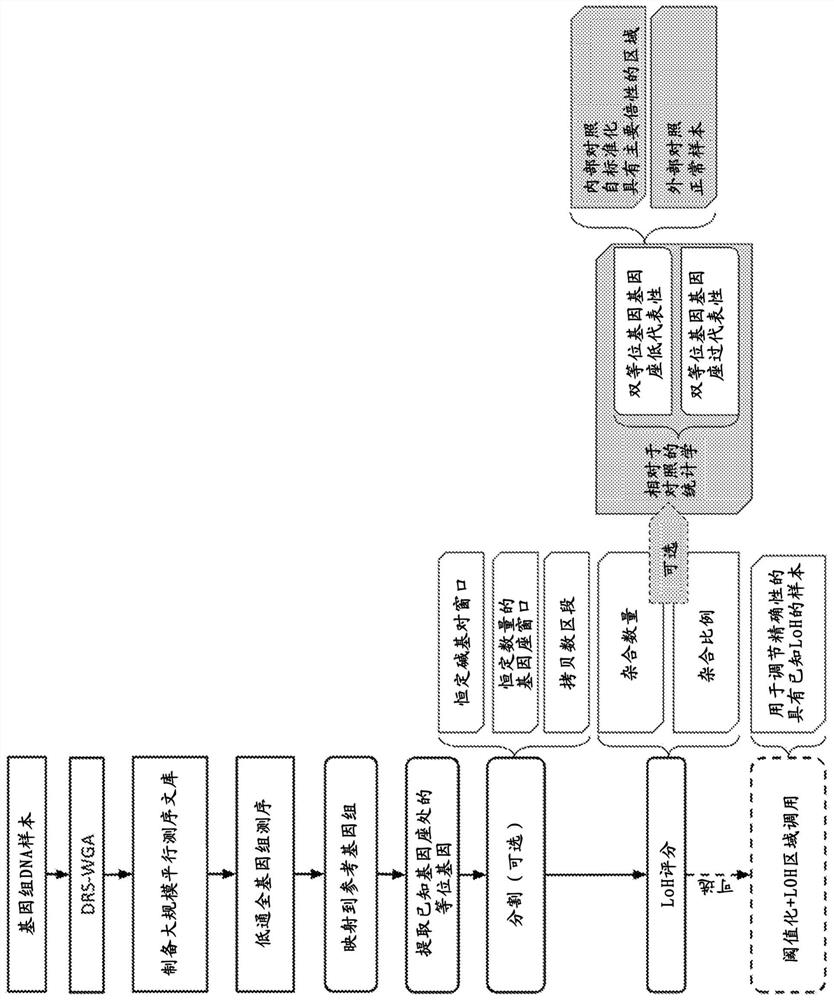
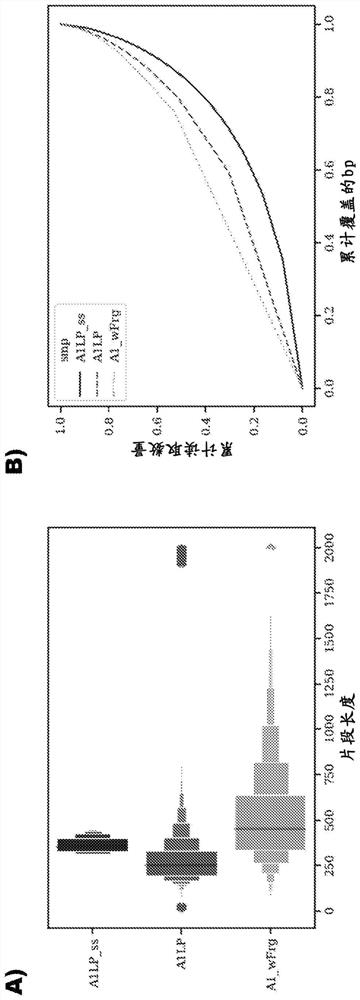
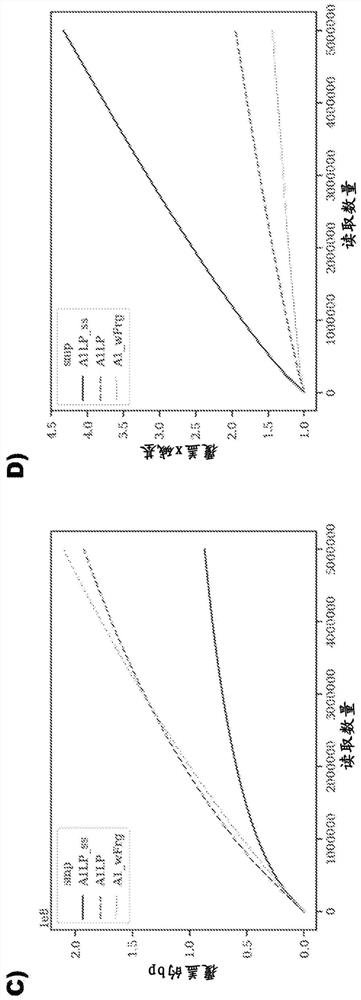
Method for analyzing heterozygosity deficiency (LoH) after genome-wide amplification of deterministic restriction site (DRS-WGA)
A technique of DRS-WGA, loss of heterozygosity, applied in the field of loss of heterozygosity in low-pass whole genome sequencing data analysis samples, which can solve the problem of insufficient information, inapplicability, and unreliable reanalysis of single cells And other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0189] In Example 1, the Ampli1 LowPass for Illumina DNA library of 1 circulating tumor cell (CTC; test) and 1 white blood cell (WBC; control) obtained from a male patient affected by multiple myeloma was considered. Sequencing reads were mapped to the hg19 reference human genome and downsampled at 1, 2, 3, 4, 5, 6, 7, 8, 9 million reads. Alleles at the dbSNP polymorphic locus (common variants of dbSNP150 with a minor allele frequency ≥ 5%) were extracted from both libraries. Loci were segmented with a fixed 10,000,000 bp genomic window. One-sided Fisher's exact test was used to assess the significance of the association between the two categories (Table 4), with the null hypothesis that the heterozygous and homozygous loci were likely to be the same in WBC (control) and CTC (test).
[0190] Table 4
[0191]
[0192] The test results for each downsampling level are as follows Figure 15 shown. Starting with 2 million reads, the method demonstrated high sensitivity in de...
Embodiment 2
[0195] In Example 2, the same single CTC data used in Example 1 was used as input and the data was downsampled at 1 million reads. In this case, the loci were partitioned in the window into a fixed number (n = 1000) of loci covered by at least 1 read. For the identification of LoH regions, the LoH score was calculated as the number of heterozygous positions in each window.
[0196] Figure 16 Detection of LoH by using a genomic window with a constant number of loci is shown. In particular, the top panel shows the copy number map of the same CTCs as in Example 1. The x-axis is chromosomes; the y-axis is copy number. Each point represents a fixed-size window of the genome. Copy number bins are indicated by solid lines. Below the plot is a heatmap representing the heterozygous counts for each genomic window. Windows with lower LoH scores (lower number of heterozygous loci) are more likely to be in LoH status, indicated by darker shades of gray. Chromosome 11, the large arm...
Embodiment 3
[0200] In Example 3, two individual Hodgkin Reed / Sternberg (Hodgkin Reed / Sternberg, HRS) cells obtained from FFPE tissue of a classic Hodgkin lymphoma sample from a male patient were analyzed for their use in Ampli1 LowPass for Illumina Libraries. Two HRS cells share the same copy number profile. Sequencing reads were mapped to the hg19 reference human genome, and alleles present at the dbSNP polymorphic locus (common variants of dbSNP150 with a minor allele frequency ≥ 5%) were extracted from both libraries. Locus partitioning using copy number segments obtained by using Control-FREEC software, enabling GC-based normalization and segmentation of copy number signals [Boeva, V et al., Bioinformatics, 27(2), 268-269 .http: / / doi.org / 10.1093 / bioinformatics / btq635). An internal control defined jointly by all regions with a copy number equal to the ploidy of the cell (copy number=2) was used. For each segment defined by copy number analysis and contained in a chromosome arm, a on...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com