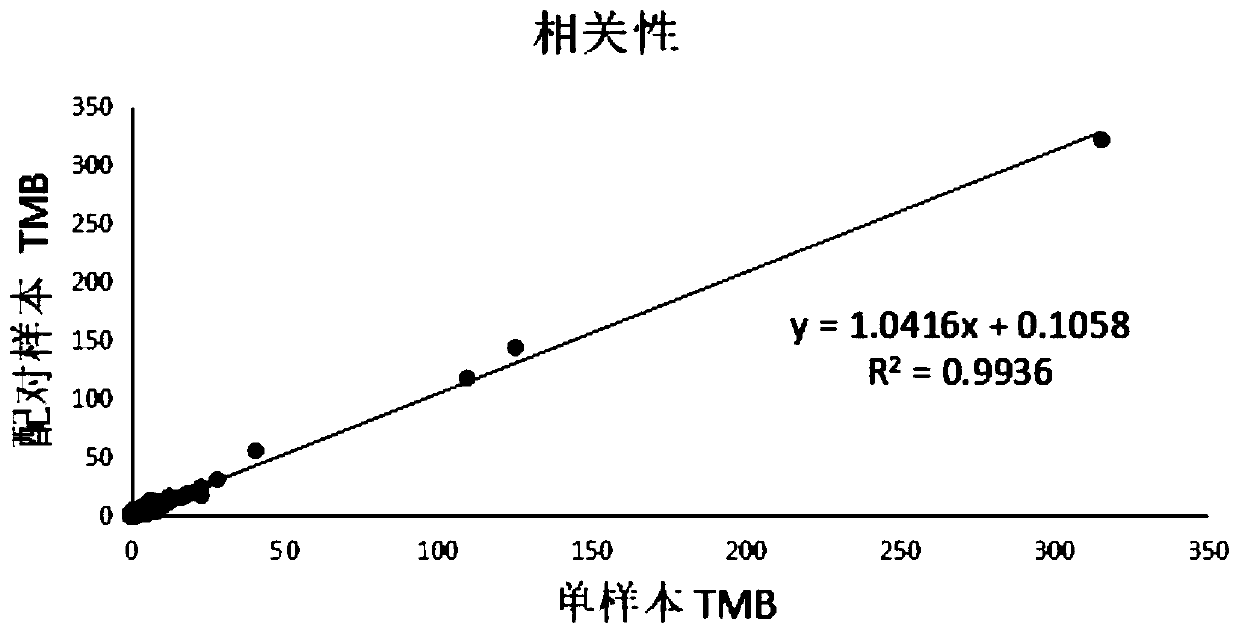
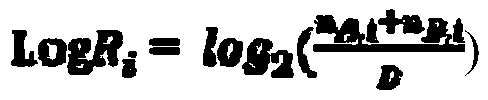Method and device for detecting tumor mutation load based on single sample
A mutation load and detection method technology, applied in the field of biomedicine, can solve the problems of large differences, the inability to make accurate descriptions of mutation backgrounds, relying on the quality and diversity of public databases, etc., to achieve accurate detection results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0089] The present invention provides a probe composition, including probes that capture all 4847 exon regions of the 312 genes shown in Table 1, and capture the introns, promoters, and fusion breakpoint regions of the 38 genes shown in Table 2 probes, and probes that capture 1778 coding regions of 709 other related genes shown in Table 3. It can be understood that the key of the present invention lies in the design of the capture region of the probe. After the capture region is determined, the specific probe design can refer to the existing capture probe design scheme, which will not be repeated here.
[0090] Table 1 All 4847 exon regions of 312 genes
[0091] ABL1 ACVR1B AKT1 AKT2 AKT3 ALK APCs AR ARAF ARID1A ARID1B ARID2 ASXL1 ATMs ATR ATRX AURKA AURKB AXIN1 AXIN2 AXL B2M BAP1 BARD1 BCL2 BCL2L1 BCOR BLM BMPR1A BRAF BRCA1 BRCA2 BRD4 BRIP1 BTK CARD11 CASP8 CBFB CBL CCND1 ...
Embodiment 2
[0099] A multigene detection kit for human tumors, including target sequence capture components, nucleic acid purification components, library construction and quality control components. Among them, the target sequence capture components include probes that capture all 4847 exon regions of the 312 genes shown in Table 1, and probes that capture the introns, promoters, and fusion breakpoint regions of the 38 genes shown in Table 2 , and probes capturing 1778 coding regions of 709 other related genes shown in Table 3. The probe composition is named as cd3 probe. As for the probe composition, conventional methods can be selected for its design. Wherein, the target sequence capturing components also include hybridization reaction solution, elution reaction solution, primers and linkers, and DNA polymerase reaction solution.
[0100] In an exemplary embodiment, the kit includes components as shown in Table 4.
[0101] Table 4 Components of Human Tumor Multigene Detection Kit
[...
Embodiment 3
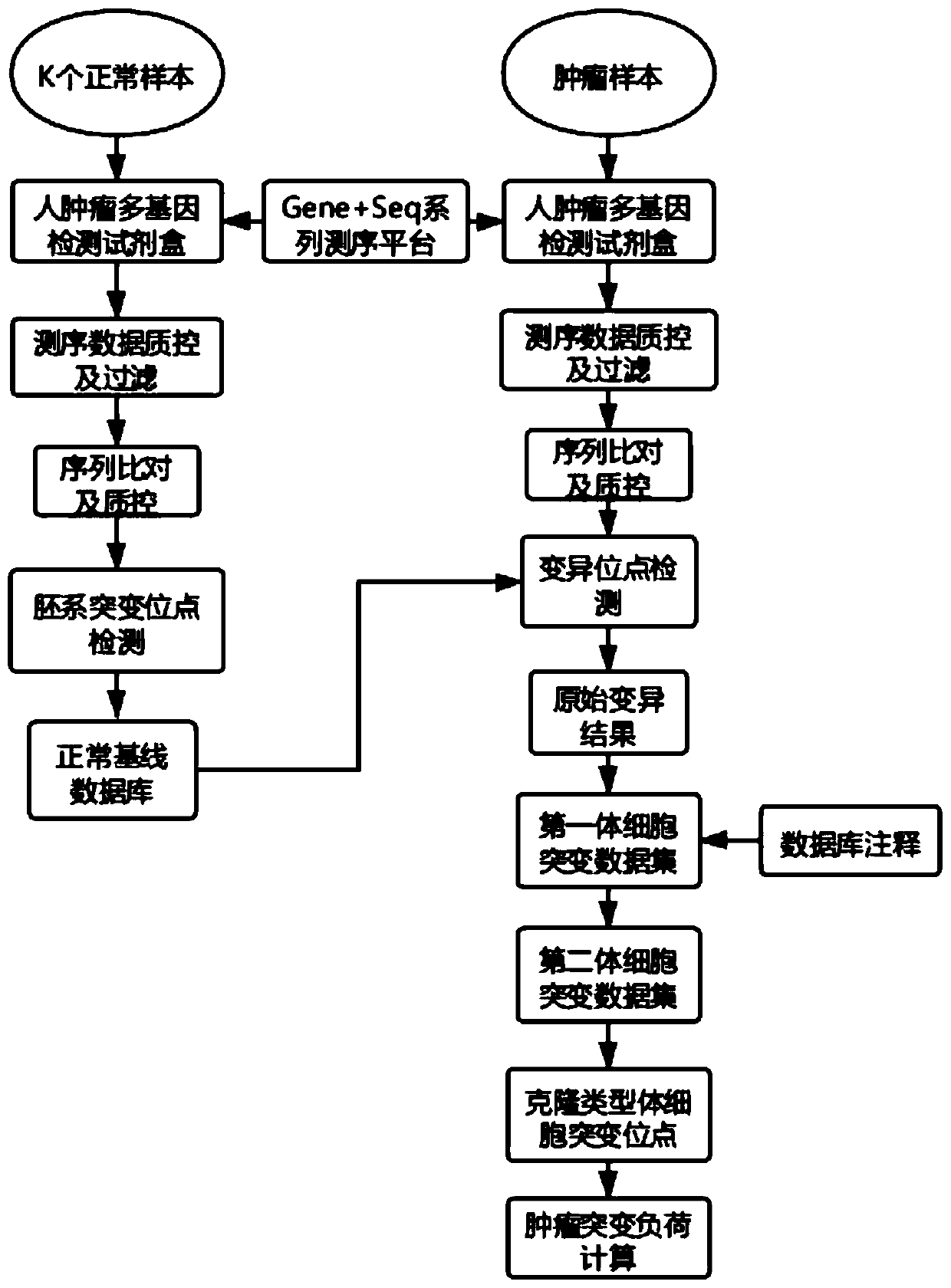
[0107] The flow chart of the single-sample-based tumor mutation burden detection method in this embodiment is as follows figure 1 shown, including the following steps:
[0108] (1) Experiment and sequencing steps
[0109] Select 241 cases of lung adenocarcinoma tumor samples, extract the DNA of the tumor samples to be tested, and use the nucleic acid purification components of the human tumor multigene detection kit (Table 4) in Example 2 to purify the DNA of the tumor samples to be tested, and use the library construction Realize library construction with quality control components, including DNA repair, DNA fragmentation, end repair and A base addition, adapter ligation and library amplification; use target sequence capture components to hybridize and capture the library, and the resulting capture products are amplified . The Gene+Seq-2000 sequencing platform was used to perform double-end sequencing with a sequence length of 100 bp to obtain the sequencing data of the tar...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com