Increasing genomic stability and reprogramming efficiency of induced pluripotent stem cells
A pluripotent stem cell, inducible technology, applied in the field of improving the genome stability and reprogramming efficiency of induced pluripotent stem cells, and can solve problems such as increased DNA damage level
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 7
[0136] Example 7 illustrates a method of generating iPSCs with improved genome stability.
[0137] Reagent test kit
[0138]Any of the compositions described herein can be included in a kit. Accordingly, kits will contain, in suitable container means, compositions related to the present invention. In particular aspects, the kit will comprise one or more agents targeting 53BP1; somatic cells; expression vectors comprising one or more agents targeting 53BP1; induced pluripotent cells produced by the methods disclosed herein ; tissues produced by induced pluripotent cells produced by the methods disclosed herein; and the like.
[0139] The components of the kit can be packaged in an aqueous medium or packaged in lyophilized form. The container means of a kit will generally comprise at least one vial, test tube, flask, bottle, syringe or other container means into which the components can be placed, and preferably aliquoted appropriately. Where more than one component is pre...
Embodiment 1
[0143] Example 1 - Materials and methods used in Examples 2-6
[0144] animal farming
[0145] To generate Bard1 mutants, heterozygous Bard1 in the C57BL / 6J background K607A / + and Bard1 S563F / + Females (Billing et al., 2018) were bred to males of the same genotype. The Brca1Smarcal1 genotype pool is composed of Brca1 from a mixed C57BL / 6J and 129Sv background tr / + Smarcal1 + / - resulting from hybridization between animals. The Brcaltr / + allele is described in (Ludwig et al., 2001). Mutant mice for Smarcal1 were obtained from the International Mouse Phenotyping Consortium (IMPC). The Brca153bp1 combined genotype set is composed of Brca1 in a mixed C57BL / 6J and 129Sv background tr / + 53bp1 + / - Produced by crosses between males and females. For the Abraxas Bach1 CtIP genotype pool (referred to as ABC), mixed background (C57BL / 6J and 129Sv) AABB mice were first crossed with CC to generate F1 triple heterozygous A+B+C+ animals. F1 generations of A+B+C+ were crossed to o...
Embodiment 2
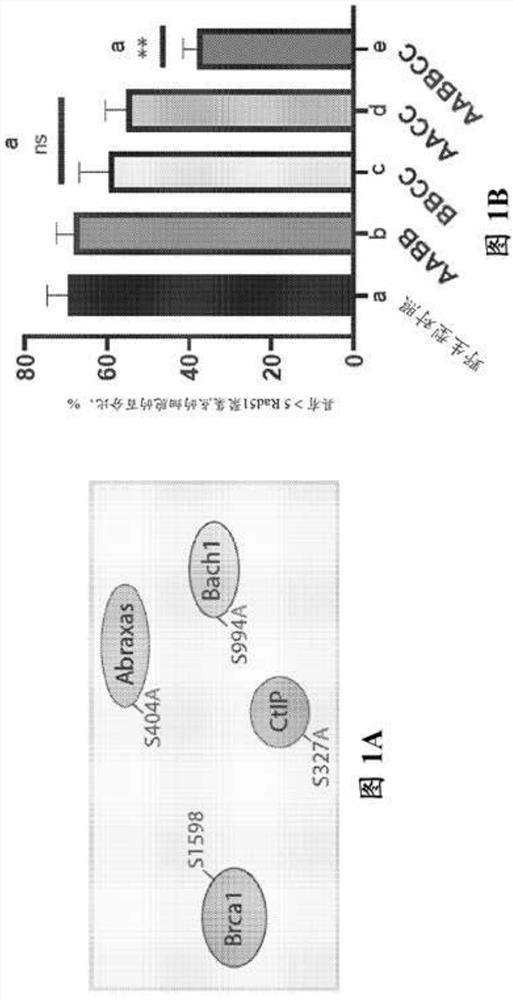
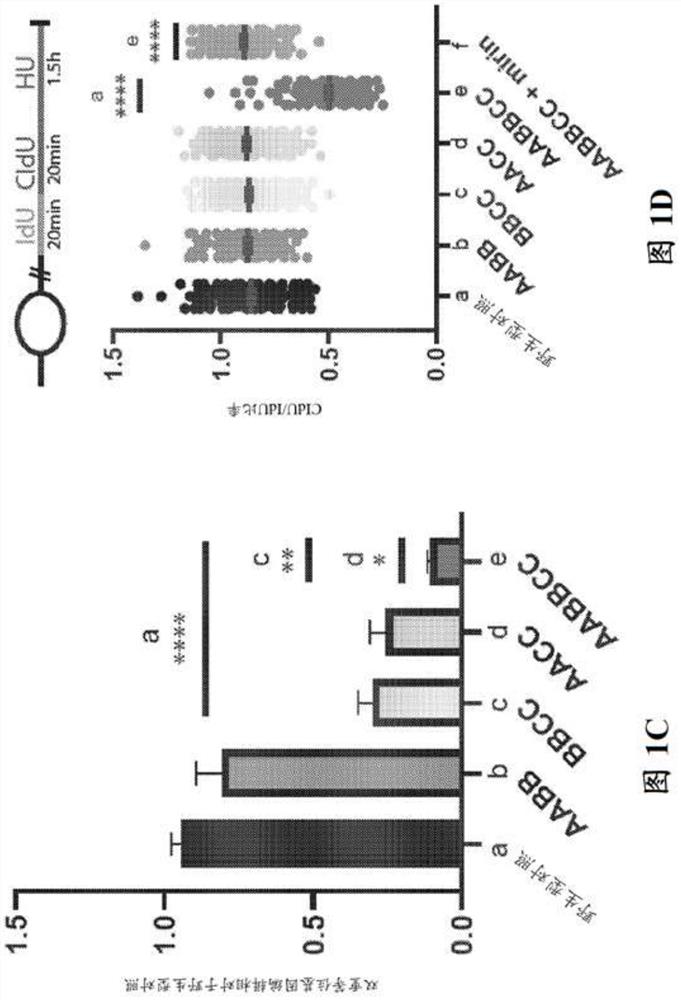
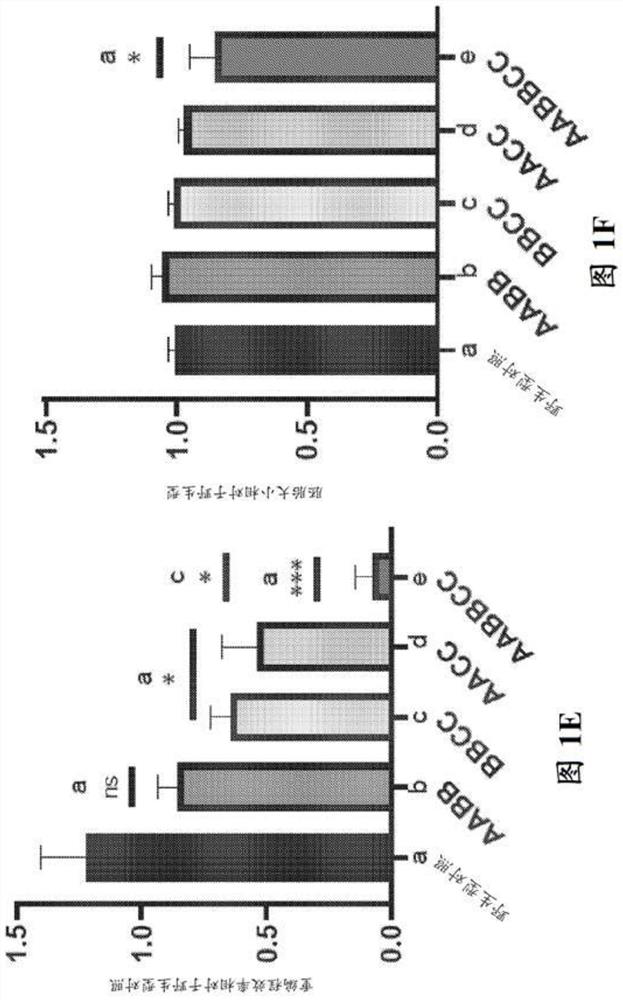
[0167] Example 2 - Reprogramming depends on the association of BRCA1 with its BRCT domain phosphorous ligands Abraxas, Bach1 and CtIP interaction
[0168] It was previously reported that impairment of Brca1 in two pathogenic Brca1 tr / tr and Brca1 S1598F / S1598F Reprogramming was severely impaired in mouse fibroblasts homozygous for either (Gonzalez et al., 2013). Although the Brca1tr allele encodes a C-terminally truncated protein lacking several key BRCA1 domains, including its SQ cluster region, PALB2-binding sequence, and BRCT motif (Ludwig et al., 2001), the protein product of Brca1S1598F Contains a missense mutation that specifically disrupts the phosphate-binding cleft of the BRCT domain (Shakya et al., 2011). Due to its BRCT phosphorous recognition domain, BRCA1 can interact in a mutually exclusive manner with phosphorylated isoforms of several DNA repair factors, including ABRAXAS, BACH1 / BRIP1 / FANCJ, and CtIP (Cantor et al., 2001; Wang et al., 2007 ; Yu, 1998). ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com