DNA methylation biomarker or composition for early liver cancer detection and application of DNA methylation biomarker or composition
A biomarker, early-stage liver cancer technology, applied in recombinant DNA technology, DNA/RNA fragments, combinatorial chemistry, etc., can solve the problems of unsatisfactory liver cancer screening effect, low detection rate and early diagnosis rate, etc. The effect of large-scale promotion and application, improving detection rate and improving prediction accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Example 1 Detection method of plasma cfDNA methylation markers for liver cancer detection
[0053] 1. Plasma cf DNA extraction: The specific operation steps of plasma cf DNA extraction are carried out in accordance with the "Operation Manual of Cell-Free DNA Extraction Kit (Suction Filtration Method)" combined with the vacuum filtration pump: the plasma is cleaved with proteinase K and lysate, and vacuum filtration is used. The pump and DNA-binding column bind the DNA and are washed with washing solution and absolute ethanol, and finally eluted with elution buffer.
[0054] 2. Fragment screening of cfDNA using magnetic beads:
[0055] ①Add 0.75 times the cfDNA product SPRIselect magnetic beads to cfDNA, vortex to mix, and incubate at room temperature for 5 minutes; place on a magnetic stand, and after the solution is clarified, pipette the supernatant into a new 1.5mL centrifuge tube for use ;
[0056] ② Add 1.05 times the SPRIselect magnetic beads of the cfDNA produc...
Embodiment 2
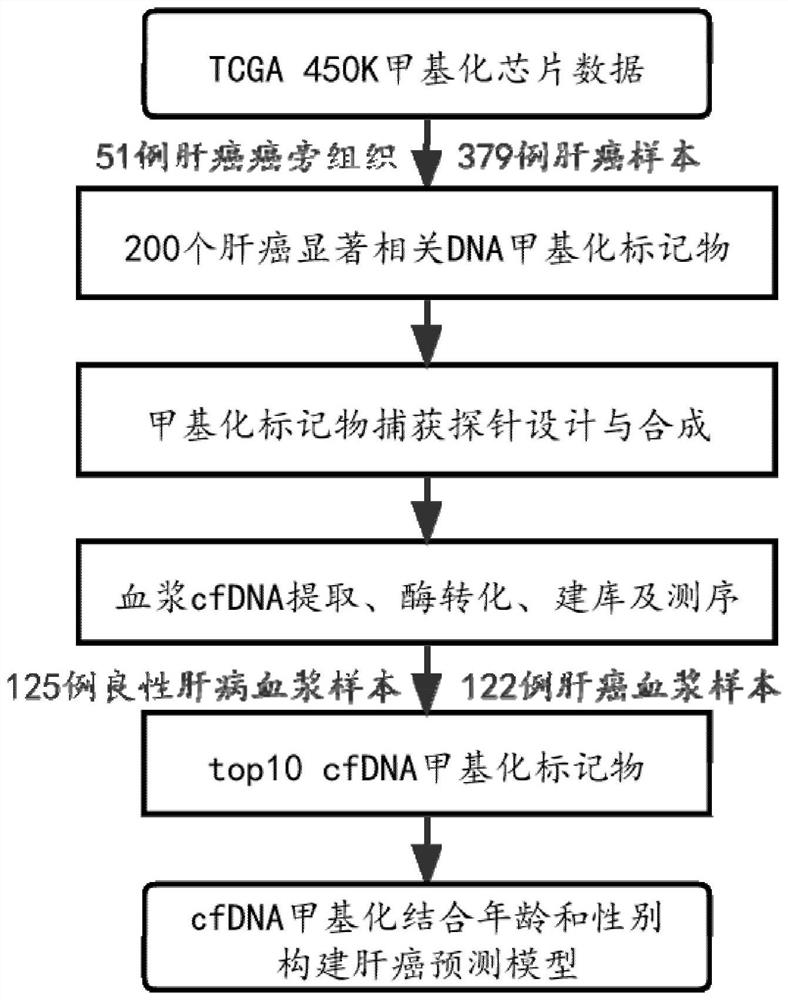
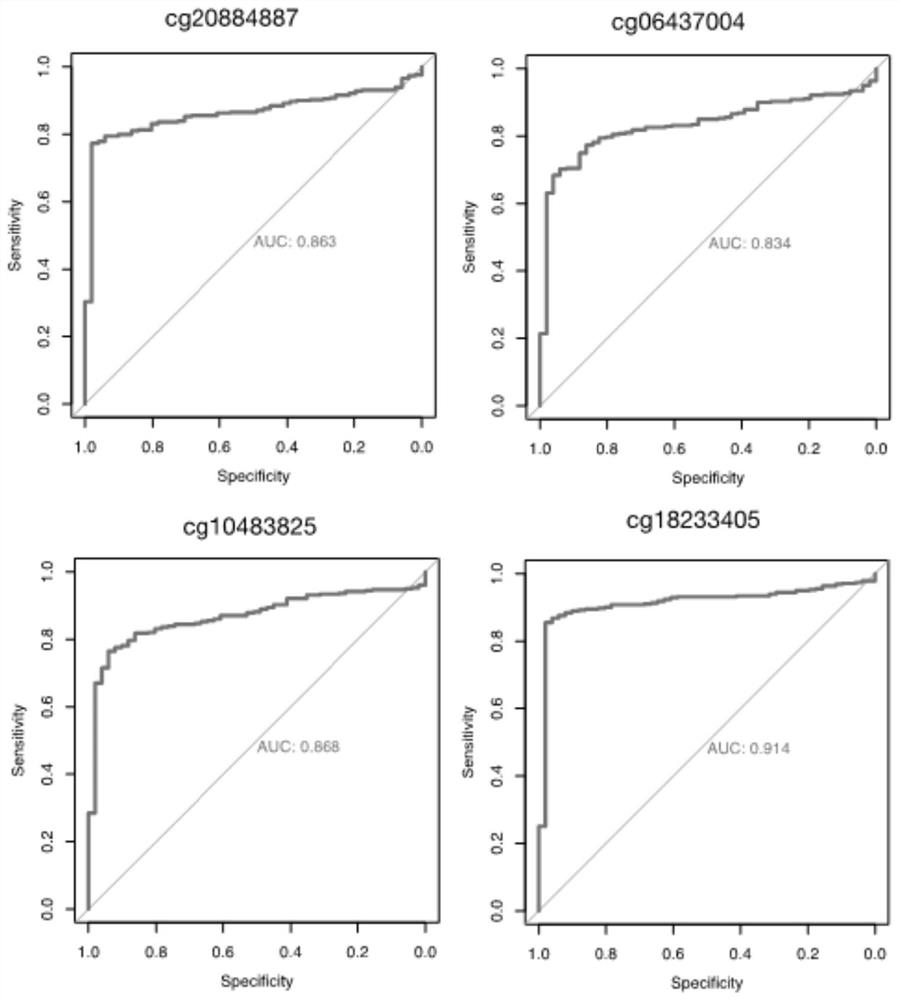
[0154] This example discloses DNA methylation specific markers for detecting liver cancer. Based on 51 liver cancer adjacent samples and 379 liver cancer samples in the TCGA public database, 10 DNA methylation sites with specific and significant methylation differences in liver cancer were mined, including 4 specific methylation sites in liver cancer. Sites with increased methylation and 6 sites with specific decreased methylation in liver cancer. These loci have high AUC values in distinguishing normal liver samples from liver cancer samples, see figure 2 and image 3 . Among them, cg18233405 and cg18842353 performed the best with AUCs of 0.914 and 0.960, respectively. Models constructed using 10 methylation biomarkers can accurately distinguish normal liver samples from early liver cancer samples, see Figure 4 .
Embodiment 3
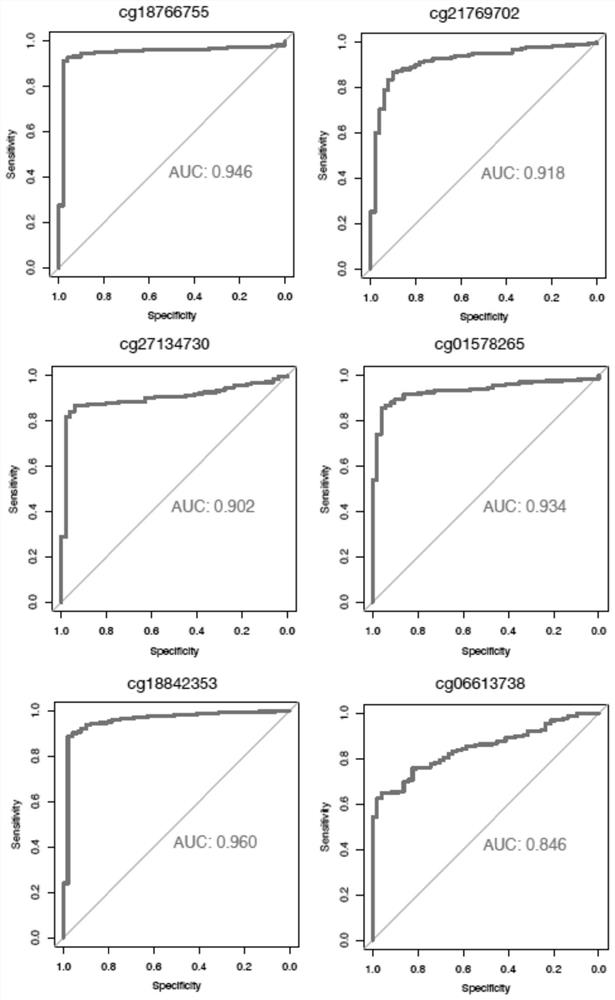
[0156] In this example, based on 125 plasma samples of benign liver disease and 122 plasma samples of liver cancer, the 10 DNA methylation markers disclosed in the present invention for detection of liver cancer were verified. The AUC values of each methylation marker independently distinguishing liver cancer plasma are shown in Figure 5 and Image 6 .
[0157] The model constructed using the combination of 4 markers with increased methylation and 6 markers with decreased methylation has an AUC value of 0.899 to distinguish benign liver disease plasma from liver cancer plasma, see Figure 7 In A, it shows that all 10 methylation markers have a certain effect on predicting liver cancer, and the model constructed together has the best predictive performance. Since studies have shown that the occurrence of liver cancer is significantly correlated with age and gender, the present invention further combines age, gender and methylation markers to construct an early-stage liver ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com