Formate dehydrogenase mutant and application thereof
A technology for formate dehydrogenase and mutants, which is applied in the field of enzyme engineering, can solve the problems of low market competitiveness, limited application, low synthesis activity, etc., and achieves the effects of market competitiveness and production cost reduction.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1 Preparation of formate dehydrogenase mutants
[0047] The present embodiment provides a formate dehydrogenase, and its preparation method is as follows:
[0048] 1. Preparation of Recombinant Plasmids and Recombinant Bacteria
[0049] The parent formate dehydrogenase used in this example is from Pandoraea commovens, and its amino acid sequence is shown below (GenBank: VVE30574.1):
[0050] MAKIVCVLYDDPVTGYPKTYARDDLPKIECYPDGQTLPTPRAIDFQPGALLGSVSGELGLRKYLESNGHELVVTSSKDGDNSVLDRELADAEIVISQPFWPAYMTAERIKRAKKLKMIVTAGIGSDHTDLQAAMEHGITVAEVTYCNSNSVAEHVMMTTLALVRNYLPSYQWVLKGGWNIADCVERSYDLEGMHVGTVAAGRIGLRVLRLMKPFGTHLHYLDRHRLPESVEKELNLTHHTSLESLAKVCDVVTLNCPLHPETEHMINADSLKHFKRGAYLINTARGKLCDRDAVAAALESGQLAGYGGDVWFPQPAPADHPWRSMPHHGMTPHISGTSLSAQTRYAAGTREILECYFENRPIRNEYLIVQNGKLAGVGAHSYSAGNATGGSEEAARFKKSA(SEQ ID No.1)。
[0051] The nucleotide sequence encoding this formate dehydrogenase is shown below:
[0052]ATGGCCAAGATTGTTTGTGTACTGTACGACGACCCCGTTACCGGCTACCCGAAGACCTACGCCCGCGACG...
Embodiment 2
[0083] Example 2 Enzymatic activity assay
[0084] A reaction solution with a final concentration of 60 mM NMN, 150 mM formic acid, and 100 mM Tris buffer was prepared, and the pH was adjusted to 8.0. Take 4 reaction solutions (900 μl each), add 100 μl of parental PcFDH with the same protein concentration and the supernatant crude enzyme solution of 3 mutant PcFDHs respectively, react at 30°C for 10 min, and then add 100 μl of 25% trichloroacetic acid to stop the reaction, The NMNH content in the reaction solution was measured by HPLC, and the specific activity of each enzyme was calculated, and the enzymatic activity of converting 1 nmol of NMN to NMNH within 1 minute was defined as 1U. The enzymatic activities of mutant PcFDH were compared, and the results are shown in Table 3.
[0085] Table 3 Relative enzyme activity detection results
[0086]
[0087] It can be seen from the above results that the enzyme activity of the single-site mutant provided in the examples of ...
Embodiment 3
[0088] Example 3 Preparation of NMNH
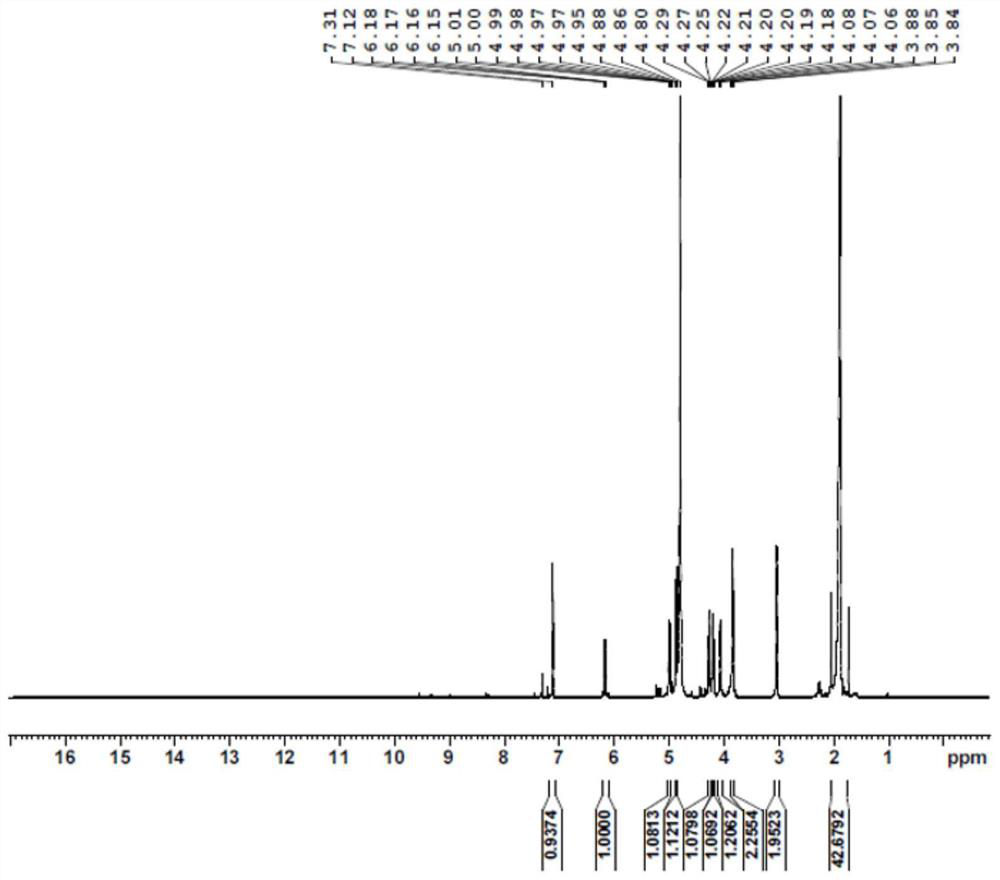
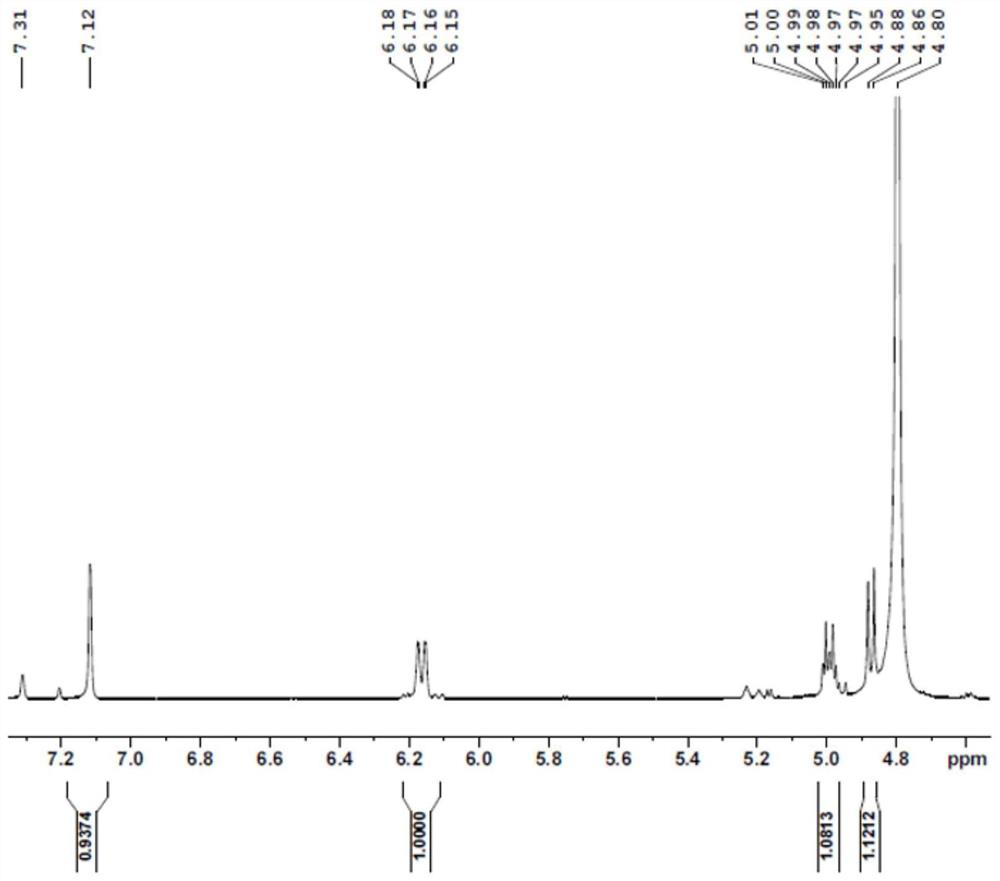
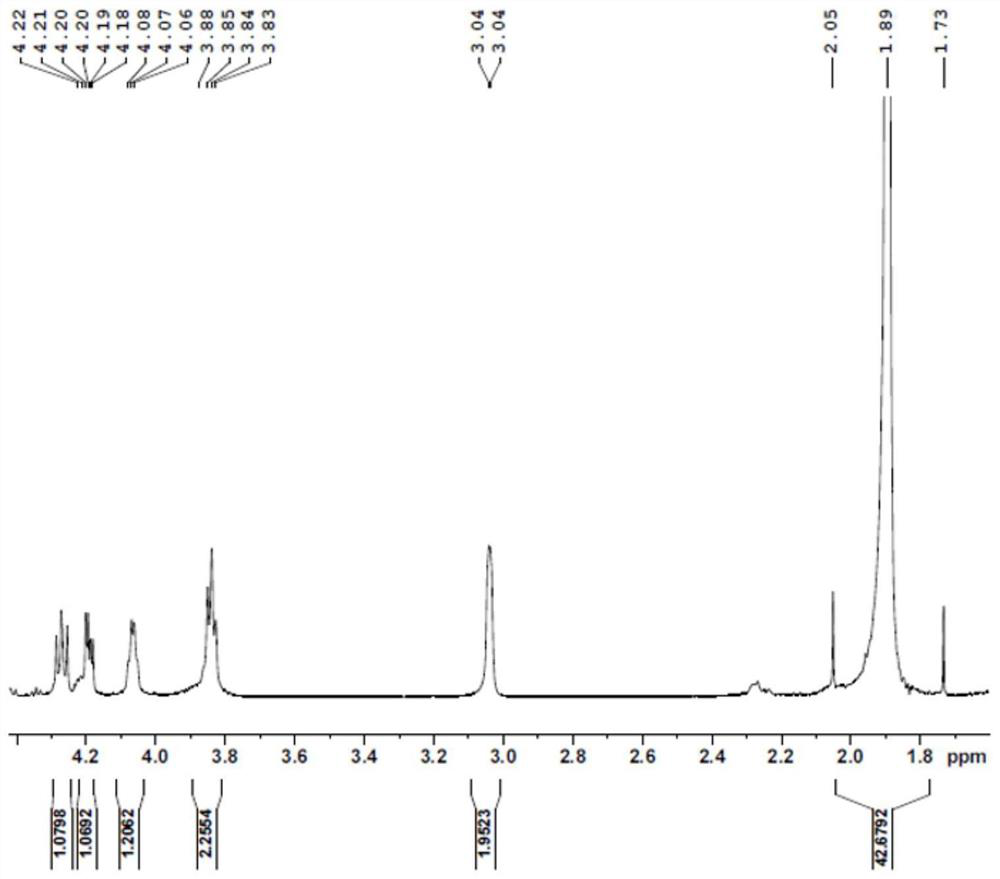
[0089] A substrate solution containing 100 mM NMN, 200 mM formic acid, and 200 mM Tris-HCl buffer was added to the reactor, and the pH was adjusted to 7.0-8.0. Then, the catalytic enzyme was added in an amount of 20ml / L (crude enzyme solution / substrate solution) of the supernatant crude enzyme solution of the mutant V54N / A121K / E228K, and after stirring evenly, the reaction was carried out in a constant temperature water bath shaker. The rotating speed of the shaker was set at 50 rpm, the reaction temperature was controlled at 30 °C, and the pH was maintained at 7.0-8.0. After 4 hours of reaction, the solution containing the crude product was obtained, and the pre-reaction substrate solution and the post-reaction reaction solution were detected by HPLC ( Figure 1-Figure 2 ), filtered, purified and dried to obtain the final product, hydrogen spectrum ( Figure 3-Figure 5 ), carbon spectrum ( Figure 6-Figure 7 ) test confirmed it to be ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com