LncRNA XIST gene knock-in animal model construction method based on RNA targeted binding site
A technology targeting binding sites and RNA targeting, applied in the field of LncRNAXIST gene knock-in animal model construction, can solve the problem of high gene off-target rate, achieve the effect of reducing the off-target rate and improving the success rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0034] 1. A method for constructing a LncRNA XIST gene knock-in animal model based on RNA targeting binding sites:
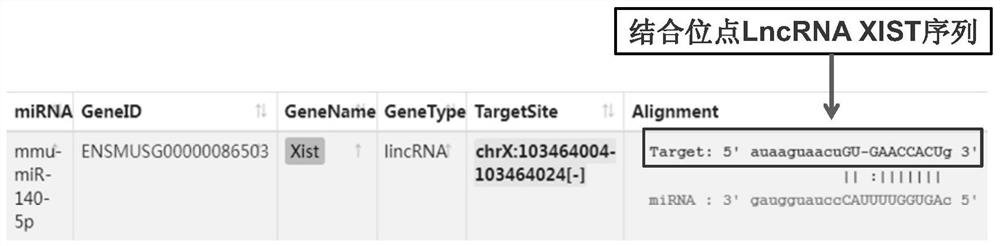
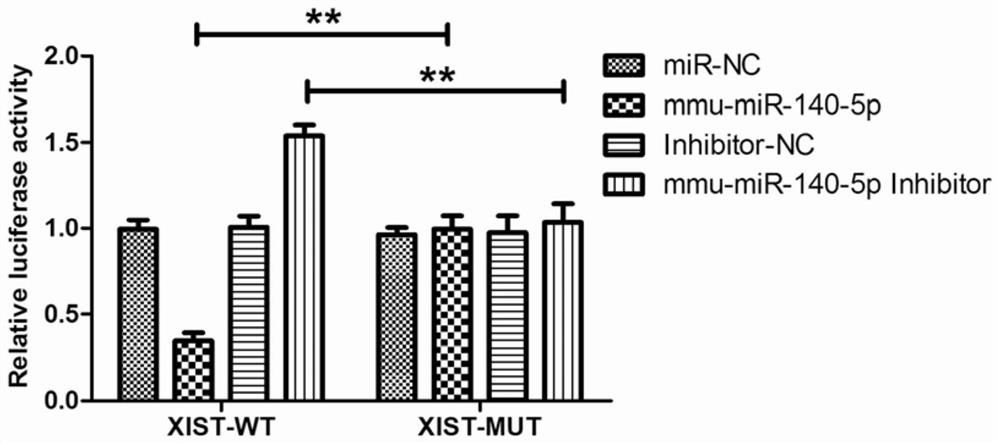
[0035] 1) Use the starbase database to perform bioinformatics analysis to confirm the targeted binding site information of LncRNA XIST and microRNA mmu-miR-140-5p, specifically LncRNAs (such as LncRNA XIST) with sequences longer than 10kb and containing multiple exons (such as LncRNA XIST) and microRNAs (such as mmu-miR-140-5p) were confirmed to have targeted binding sites in bioinformatics analysis and in vitro experiments;
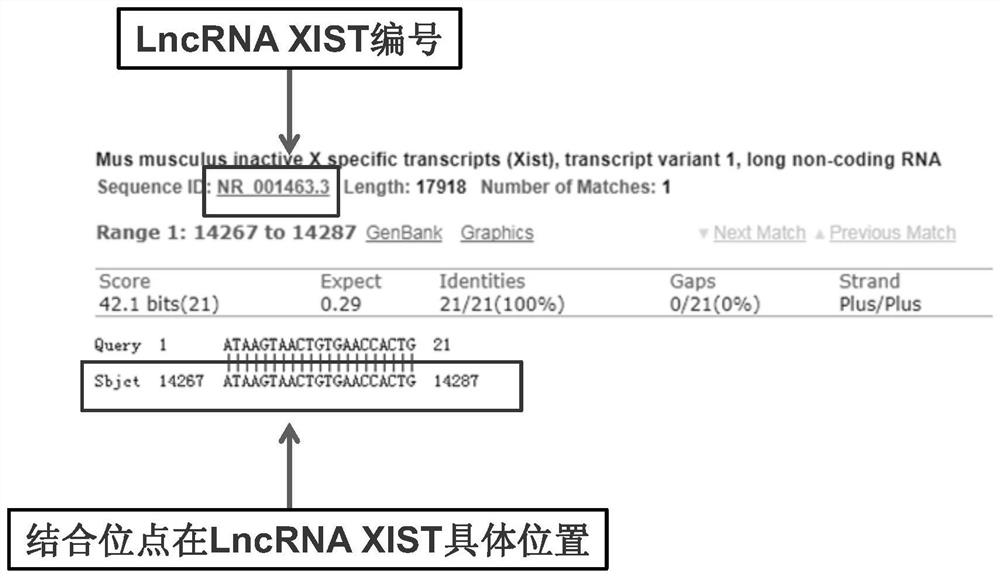
[0036] 2) Compare the position of the above-mentioned targeted binding sites in the LncRNA XIST gene through the NCBI database, specifically, LncRNA XIST (length 17.9kb) and mmu-miR-140-5p have been confirmed to have targeting in bioinformatics analysis and in vitro experiments. binding site;
[0037] 3) Compare the above-mentioned targeted binding site in the seventh exon of the LncRNA XIST gene through the NCBI database;
[0038] 4) Th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com