Oligonucleotide primer for effective detection of hepatitis C virus (HCV) and its use
A hepatitis C virus and oligonucleotide technology, applied in the field of sequence improvement, can solve the problems of indistinguishable infection, undetected HCV infection, and unsuccessful development
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
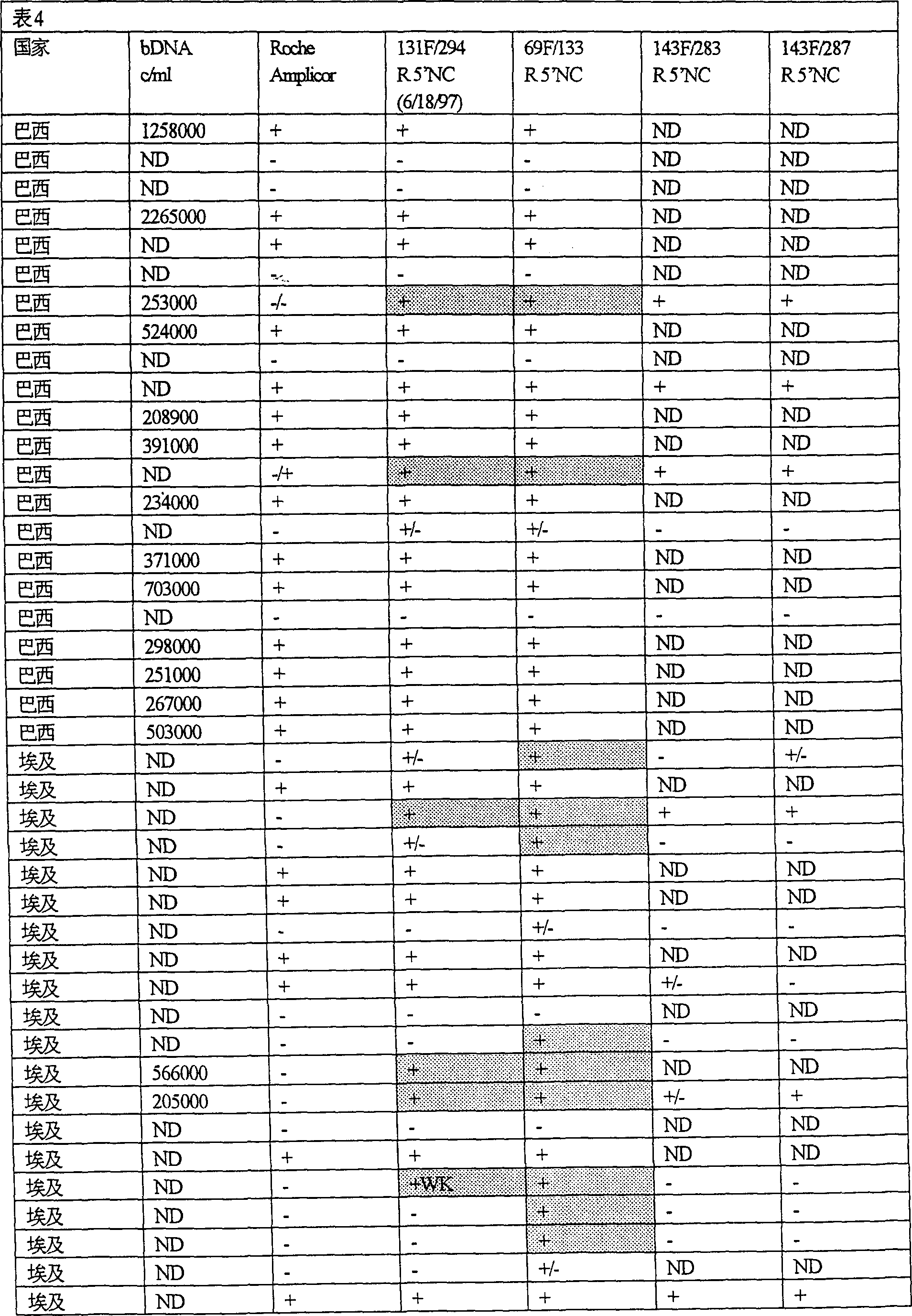
[0144] Example 1: Detection of HCV in Biological Samples: Comparison of 5' HCV Amplification Primers and Roche Amplicor HCV Identification.
[0145] The following experiments were performed to compare the HCV identification of the present invention and the Roche Amplicor system.
[0146] A method:
[0147] 1. Sample preparation:
[0148] RNA was prepared from plasma samples using Pure Script RNA Isolation Reagent (Gentra Systems, Minneapolis MN). Modifications to the manufacturer's protocol for body fluid preparation included using 40 g of glycogen instead of 20 g of glycogen as a carrier to aid in the precipitation of viral RNA. Also, in most cases, after precipitating RNA with isopropanol and washing the RNA pellet with ethanol, the RNA pellet was resuspended in RT buffer mix rather than in the RNA hydration solution provided by the manufacturer.
[0149] 2. Reverse transcription
[0150] Add 100u of recombinant Moloney murine leukemia virus (M-MLV) reverse ...
Embodiment 2
[0176] Embodiment 2: Analysis of the sensitivity of 5' NCR HCV detection method.
[0177] Use the 'NCR primer pair of the present invention to carry out the following experiments to check the sensitivity of the HCV detection method.
[0178] 1. Three patient samples were diluted according to the theoretical copy number of HCV RNA and analyzed for HCV RNA using the Roche Amplicor system and the primer pairs of the present invention. The results are shown in Table 9 below:
[0179] Table 9
theoretical copy
number / ml
Roche
Amplicor
5'NC
131F / 294R
JJCD Password
A1
500
+
+
A2
100
+
+
A3
10
-
+
A4
5
-
+
A5
1
-
-
B1
500
+
+
B2
100
+
+
B3
10
+
+
B4
5
+
+
B5
1
+
+
C1 ...
Embodiment 3
[0185] Example 3: Detection of HCV in Biological Samples: Comparison of 3' NCR HCV Amplification Primers and Roche Amplicor HCV Assay.
[0186] 150 plasma samples from Brazil that had previously tested positive for HCV antibodies were identified by Roche Amplicor and examined with the 3'NCR primers of the present invention. Reverse transcription and amplification were performed as described in Example 1. Reverse transcription was performed with primers whose sequence was 5'GTATCAGCACTC-3', and amplification was performed with X1F27 / 57R27 primer pair. Each reaction contained an internal positive control (IPC) of plasmid DNA. The amplified product was detected with a capture probe 3X30PRB25 using the Sure Cell system. The product of each reaction was run on a gel to confirm the results. Negative controls (interspersed with clinical samples) containing negative plasma samples and water (no RNA) were both involved and were negative in RT-PCR. A comparison of the results ob...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com