Method and apparatus for identifying, classifying, or quantifying DNA sequences in a sample without sequencing
a technology of dna sequences and methods, applied in the field of dna sequence classification, identification or determination, and quantification, can solve the problems of cumbersome and uneconomical adaptation of these methods to the problem of recognizing all sequences in a sample, application requiring such a large number of probes is clearly too cumbersome to be economic or practical, and cannot be applied to mixtures. , to achieve the effect of sufficient discrimination and resolution, rapid and economical, quantitative and
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
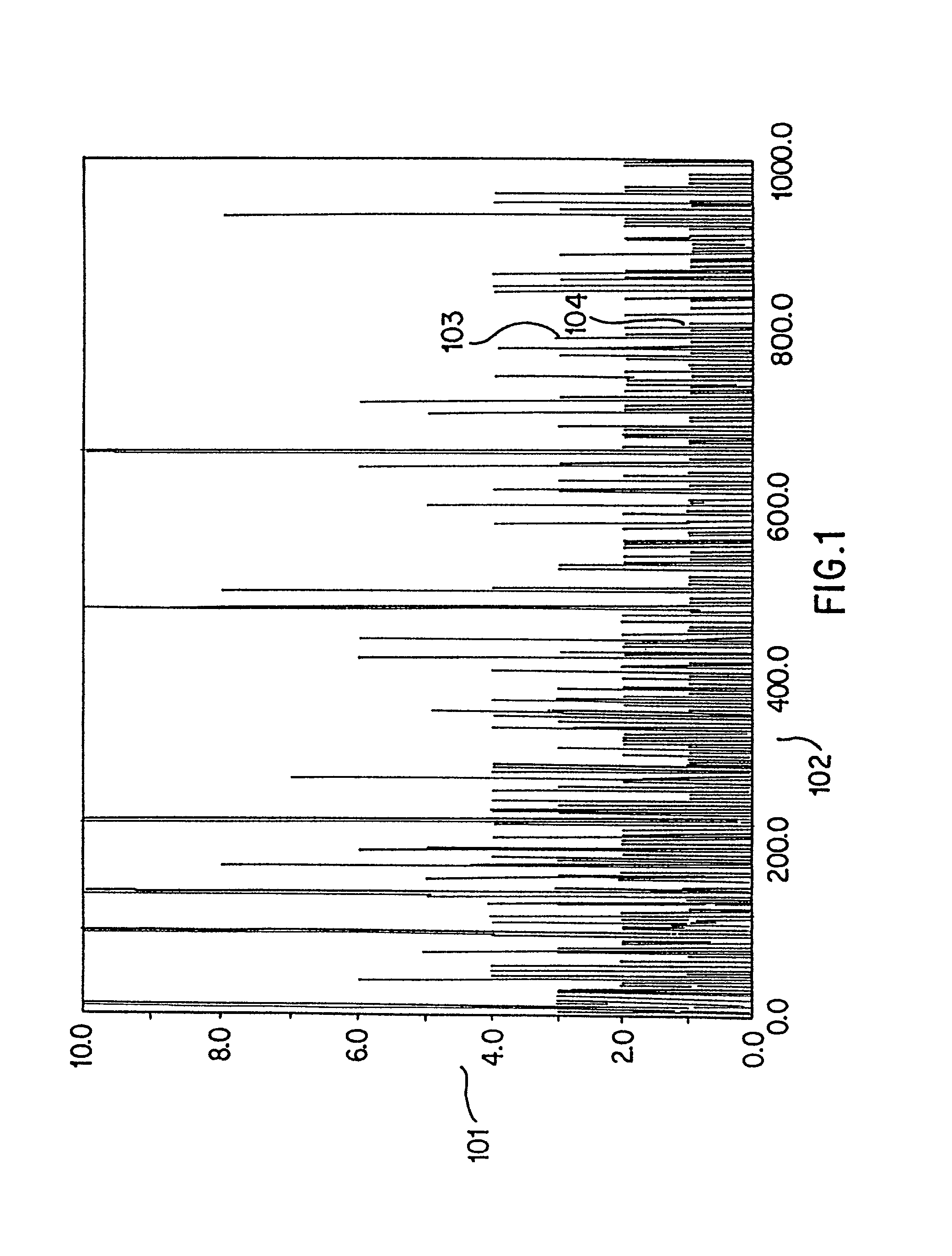
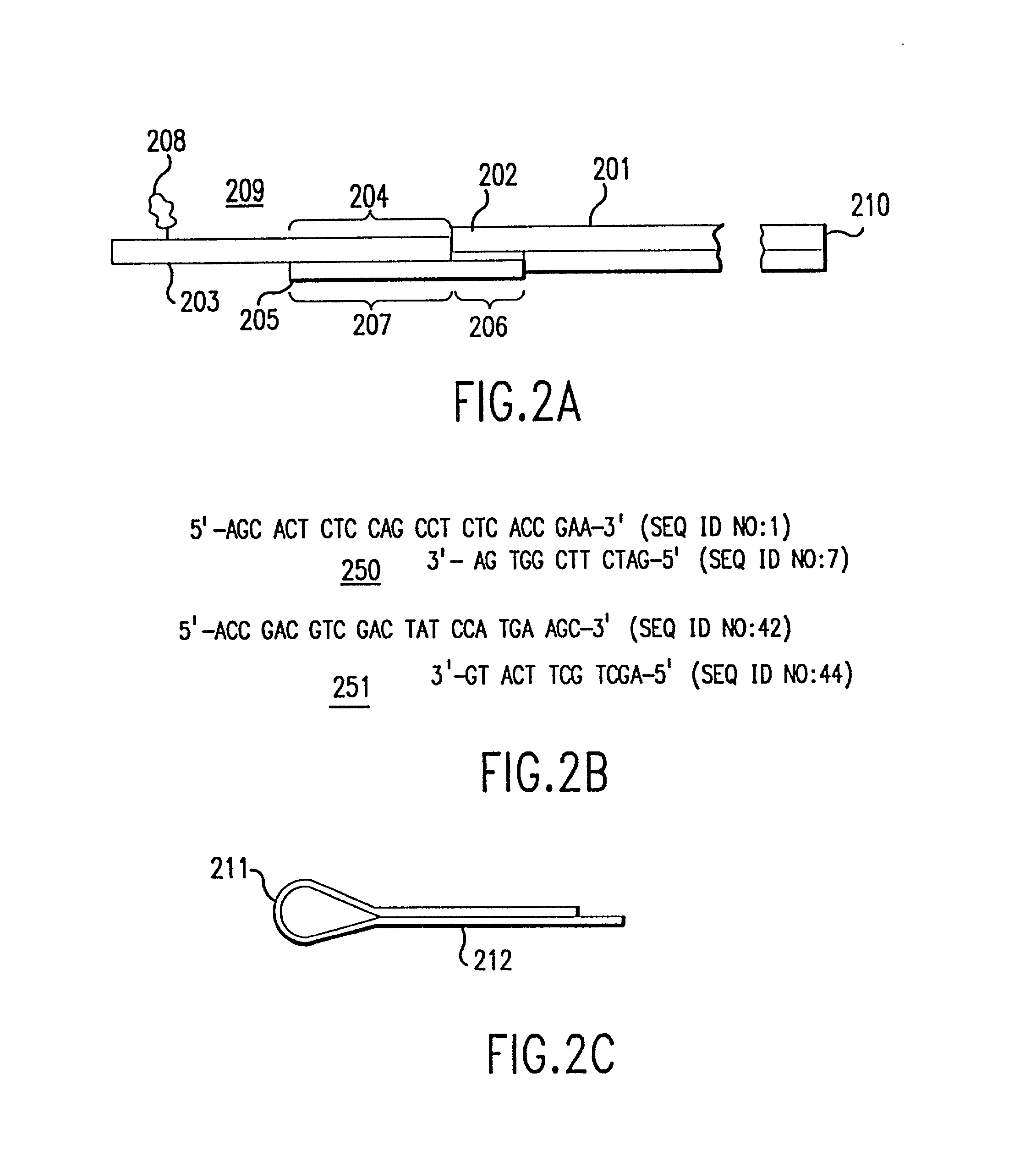
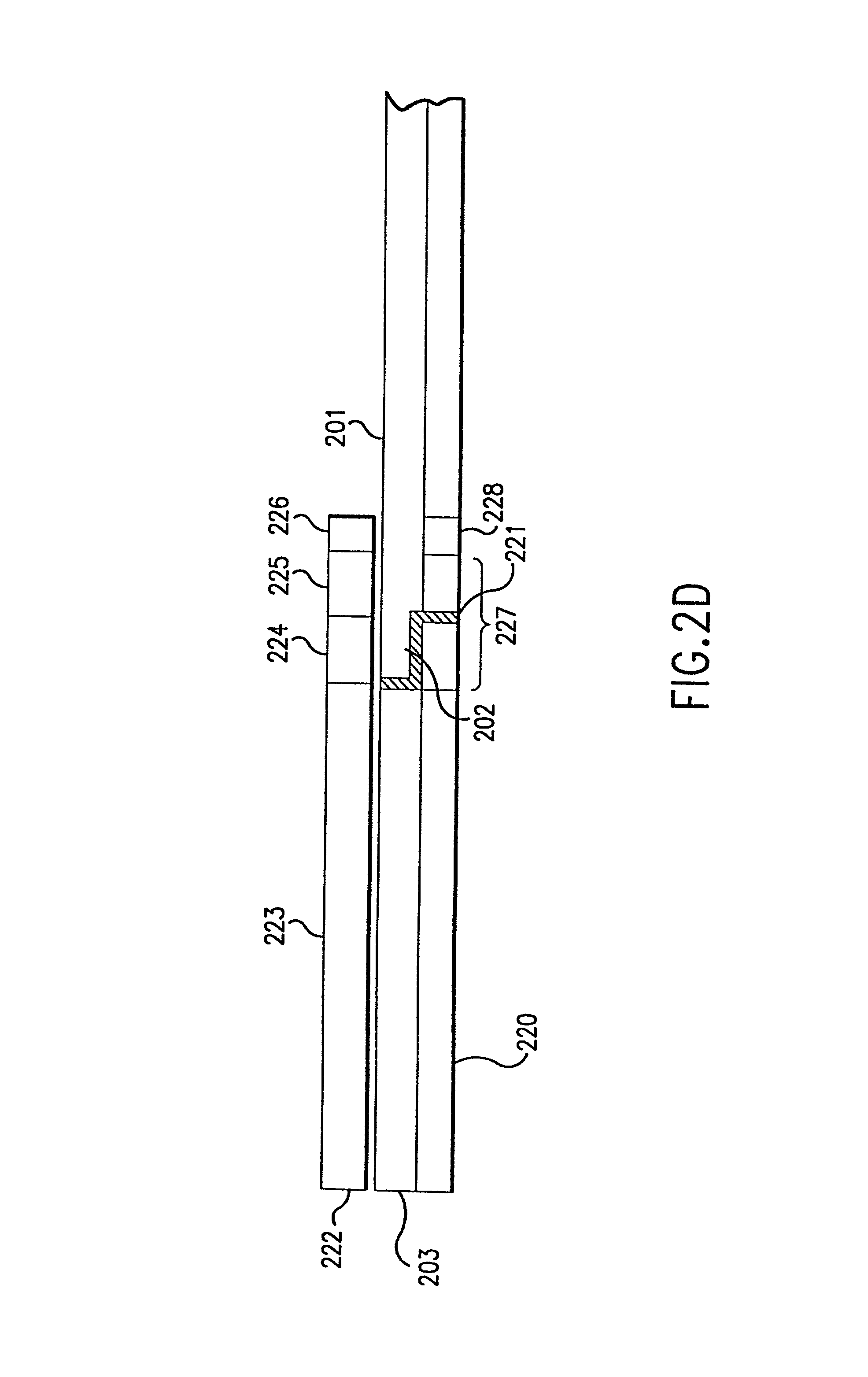
[0146] According to the present invention, to uniquely identify an expressed gene sequence, full or partial, and many components of genomic DNA it is not necessary to determine actual, complete nucleotide sequences of samples. Full sequences provide far more information than is needed to merely classify or determine a gene according to the invention. For example, in the human genome, it is known that there are approximately 10.sup.5 expressed genes. Since the average length of a coding sequence is approximately 2000 nucleotides, the total number of possible sequences is approximately 4.sup.2000, or about 10.sup.1200. The actual number of expressed human genes is an unimaginably small fraction (10.sup.-1195) of the total number of possible DNA sequences. Even sequencing a 50 bp fragment of a cDNA sequence generates about 10.sup.25 times more information than is needed for classification of that sequence. Use of the present invention allows direct classification of expressed gene sequ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Angle | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com