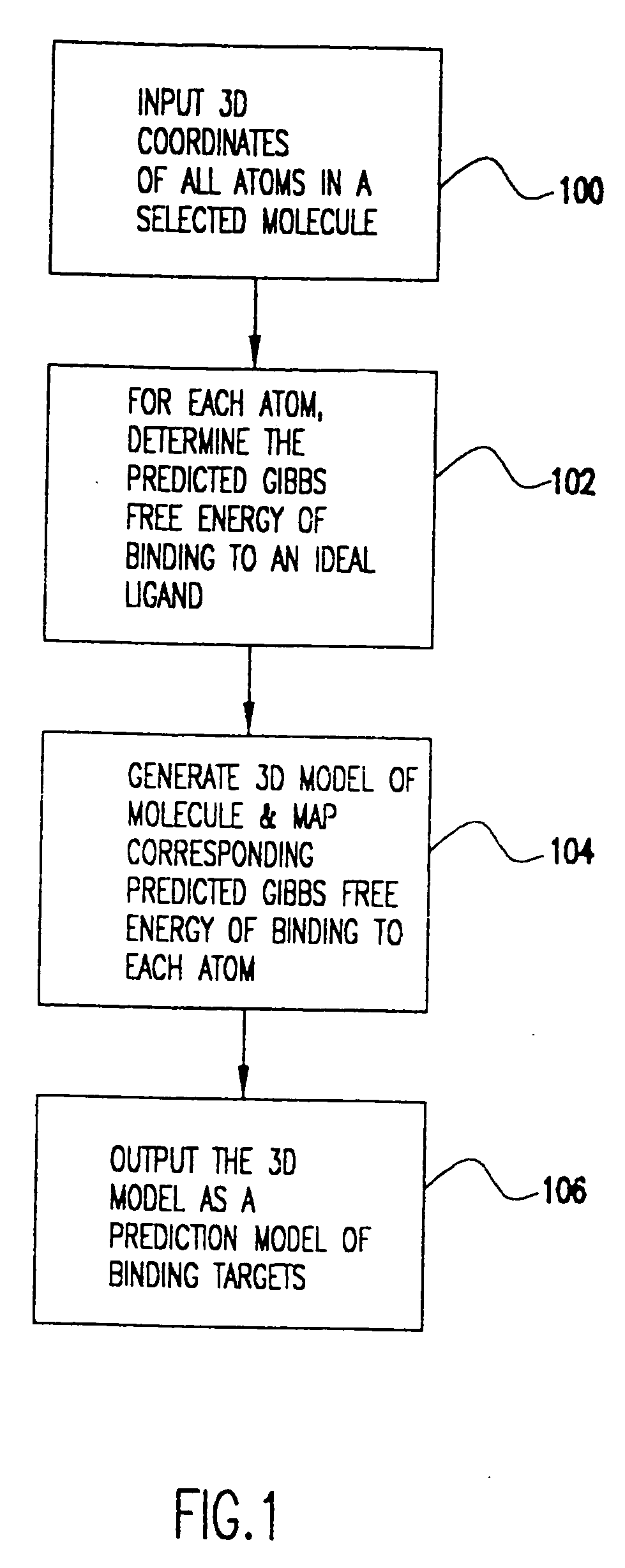
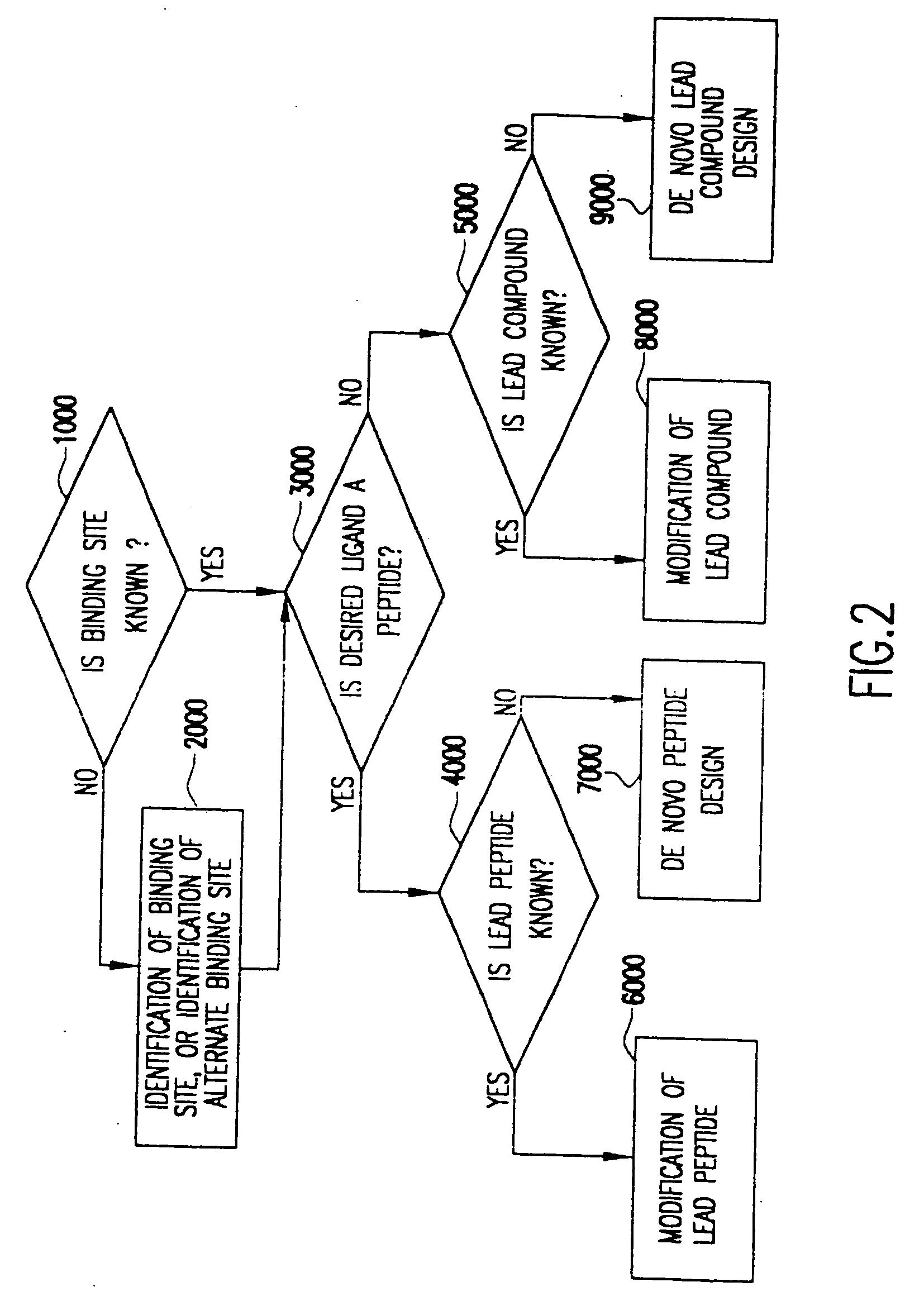
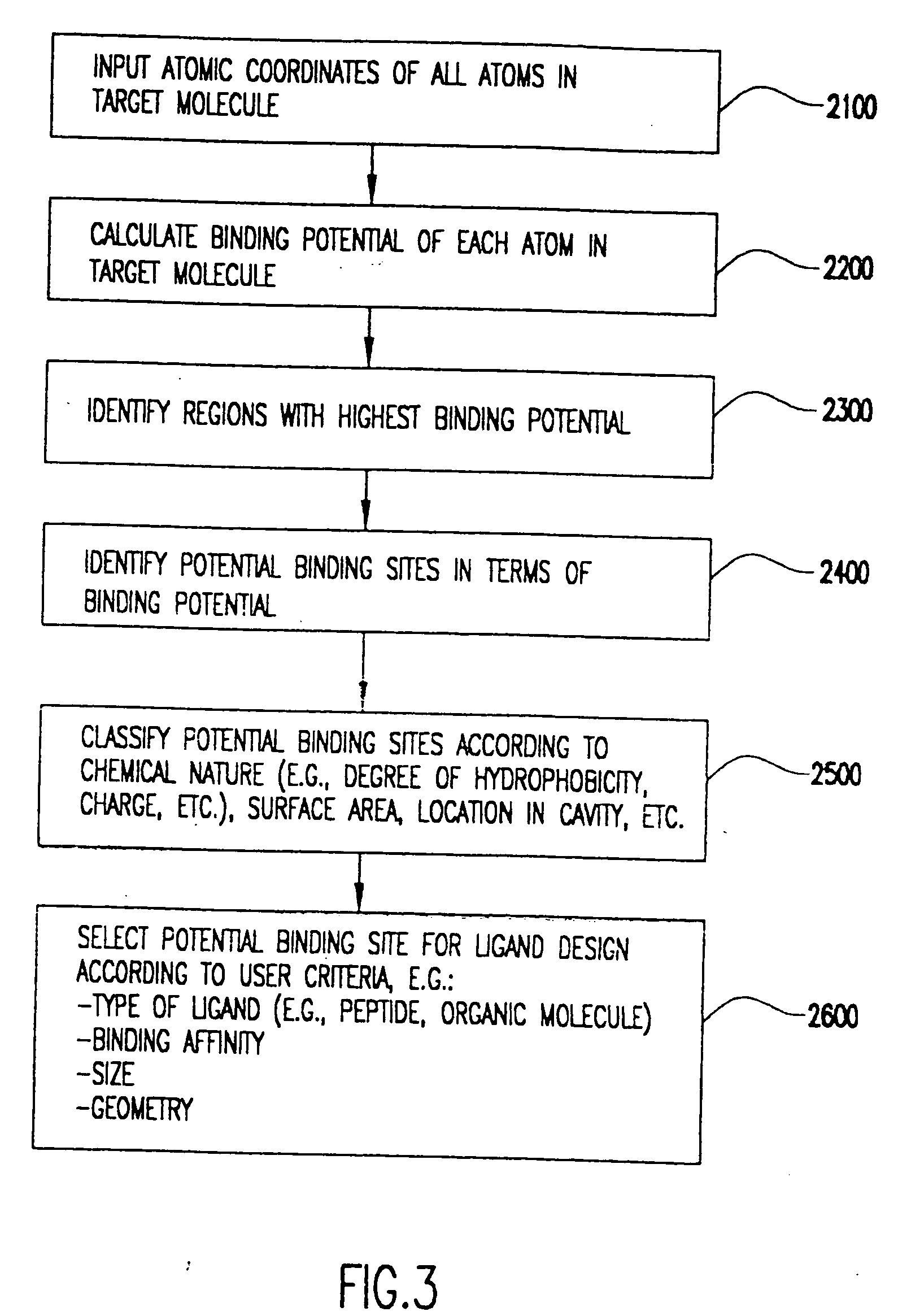
Method for the prediction of binding targets and the design of ligands
a technology for predicting binding targets and ligand design, applied in the direction of peptides, instruments, molecular structures, etc., can solve the problems of complicated situation and very demanding efforts for optimizing lead compounds
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Prediction of Binding Affinities of HIV-1 Protease Inhibitors
HIV-1 protease has been the subject of intense research during the last few years. The development of protease inhibitors is a major endeavor for several pharmaceutical companies, since the successful inhibition of this protein arrests viral maturation. Inhibitors of the HIV-1 protease are substrate analogues, i.e., they function by competing with the natural substrates for the active site. Because substrates are rapidly hydrolyzed by the protease, crystallographic structures of enzyme / substrate complexes cannot be obtained, thus creating additional obstacles to the design process.
In the example presented here, the known structure of the HIV-1 protease with the inhibitor Ace-Thr-Ile-Nle-Nle-Gln-Arg-NH2 (pdb file 4hvp) is used to generate the structure of the widely used chromogenic substrate Lys-Ala-Arg-Val-Nle-NPhe-Glu-Ala-Nle-NH2. In the chemical formulas, Nle stands for norleucine, and NPhe for p-nitro-phenylalanine...
example 2
Application of Structure-Based Thermodynamic Design of Peptide Inhibitors of the Aspartic Protease Endothiapepsin
The development of a structure parameterization of the energetics of protein folding and binding (Bardi et al., 1997; D'Aquino et al., 1996; Gomez et al., 1995(a); Gomez et al., 1995(b); Hilser et al., 1996(b); Luque et al., 1996) has been shown to be accurate enough to predict the helical propensities of amino acids with an accuracy better than 0.2 kcal / mol (Luque et al., 1996), to correctly predict the global stability of proteins and the stability constants per residue as reflected in the pattern of NMR-detected hydrogen exchange protection factors (Hilser et al., 1996(c); Hilser et al., 1997(a); Hilser et al., 1997(b)), and to predict the binding affinity of thirteen HIV-1 protease inhibitors for which high resolution structures are available with an accuracy better than ±1 kcal / mol (Bardi et al., 1997). Since the parameterization has reached the state in which accu...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Gibbs energy | aaaaa | aaaaa |
| binding affinity | aaaaa | aaaaa |
| energy minimized | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com