Use of a tag to enrich polypeptides libraries
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Immunization of Mice with Antigens and Purification of RNA from Mouse Spleens
[0146] Mice were immunized by the following method based on experience of the timing of spleen harvest for optimal recovery of mRNA coding for antibody. Two species of mice were used: Balb / c (Charles River Laboratories, Wilmington, Mass.) and A / J (Jackson Laboratories, Bar Harbor, Me.). Each of ten mice were immunized intraperitoneally with antigen using 50 μg protein in Freund's complete adjuvant on day 0, and day 28. Tests bleeds of mice were obtained through puncture of the retro-orbital sinus. If, by testing the titers, they were deemed high by ELISA using biotinylated antigen immobilized via streptavidin, the mice were boosted with 50 μg of protein on day 70, 71 and 72, with subsequent sacrifice and splenectomy on day 77. If titers of antibody were not deemed satisfactory, mice were boosted with 50 μg antigen on day 56 and a test bleed taken on day 63. If satisfactory titers were obtained, the animals...
example 2
Preparation of Complementary DNA (cDNA)
[0148] The total RNA purified as described above was used directly as template for cDNA. RNA (50 μg) was diluted to 100 μL with sterile water, and 10 μL-130 ng / μL oligo dT12 (synthesized on Applied Biosystems Model 392 DNA synthesizer at Biosite Diagnostics) was added. The sample was heated for 10 min at 70° C., then cooled on ice. 40 μL 5× first strand buffer was added (Gibco / BRL, Gaithersburg, Md.), 20 μL 0.1 M dithiothreitol (Gibco / BRL, Gaithersburg, Md.), 10 μL 20 mM deoxynucleoside triphosphates (dNTP's, Boehringer Mannheim, Indianapolis, Ind.), and 10 μL water on ice. The sample was then incubated at 37° C. for 2 min. 10 μL reverse transcriptase (Superscript™ II, Gibco / BRL, Gaithersburg, Md.) was added and incubation was continued at 37° C. for 1 hr. The cDNA products were used directly for polymerase chain reaction (PCR).
example 3
Amplification of cDNA by PCR
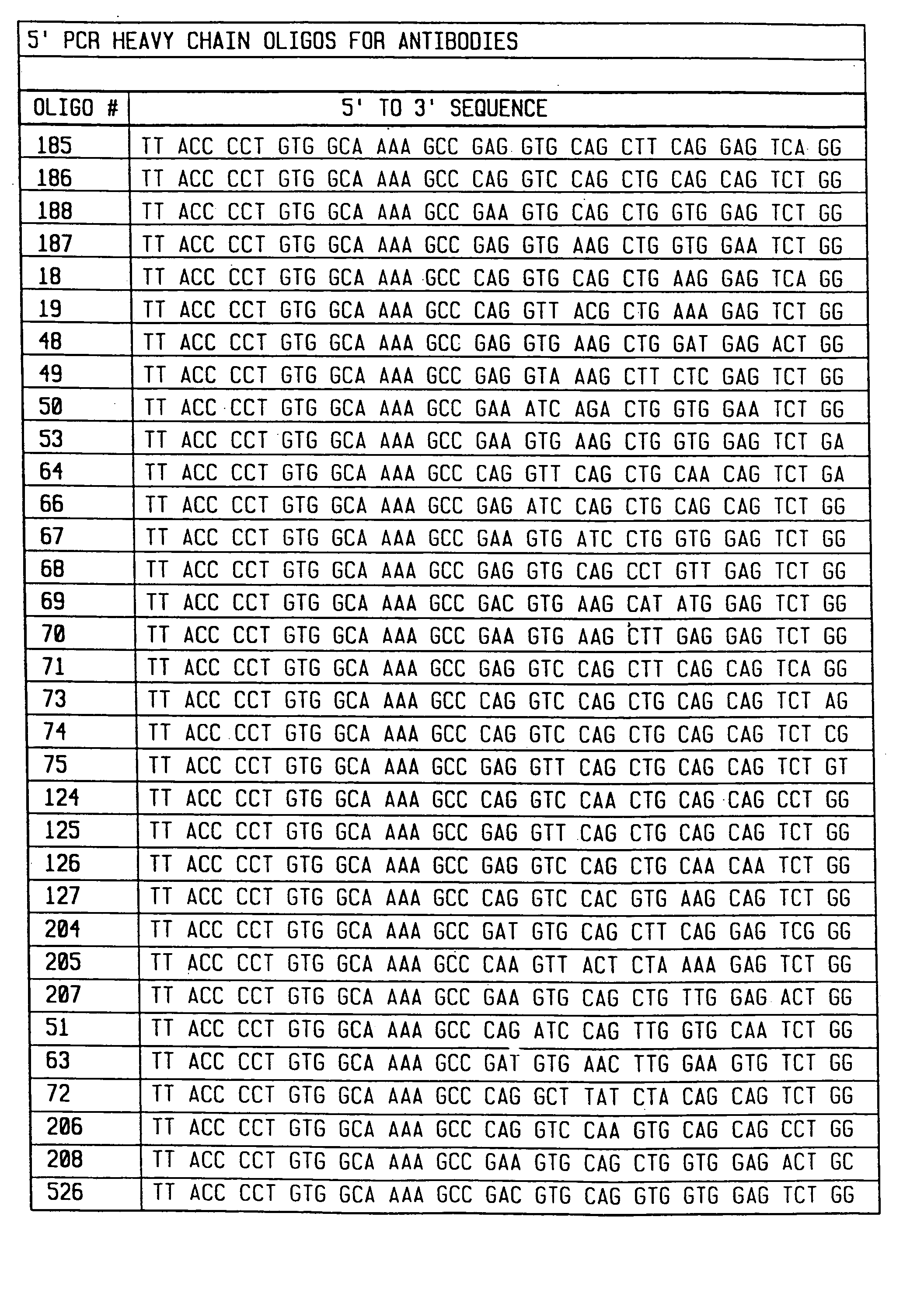
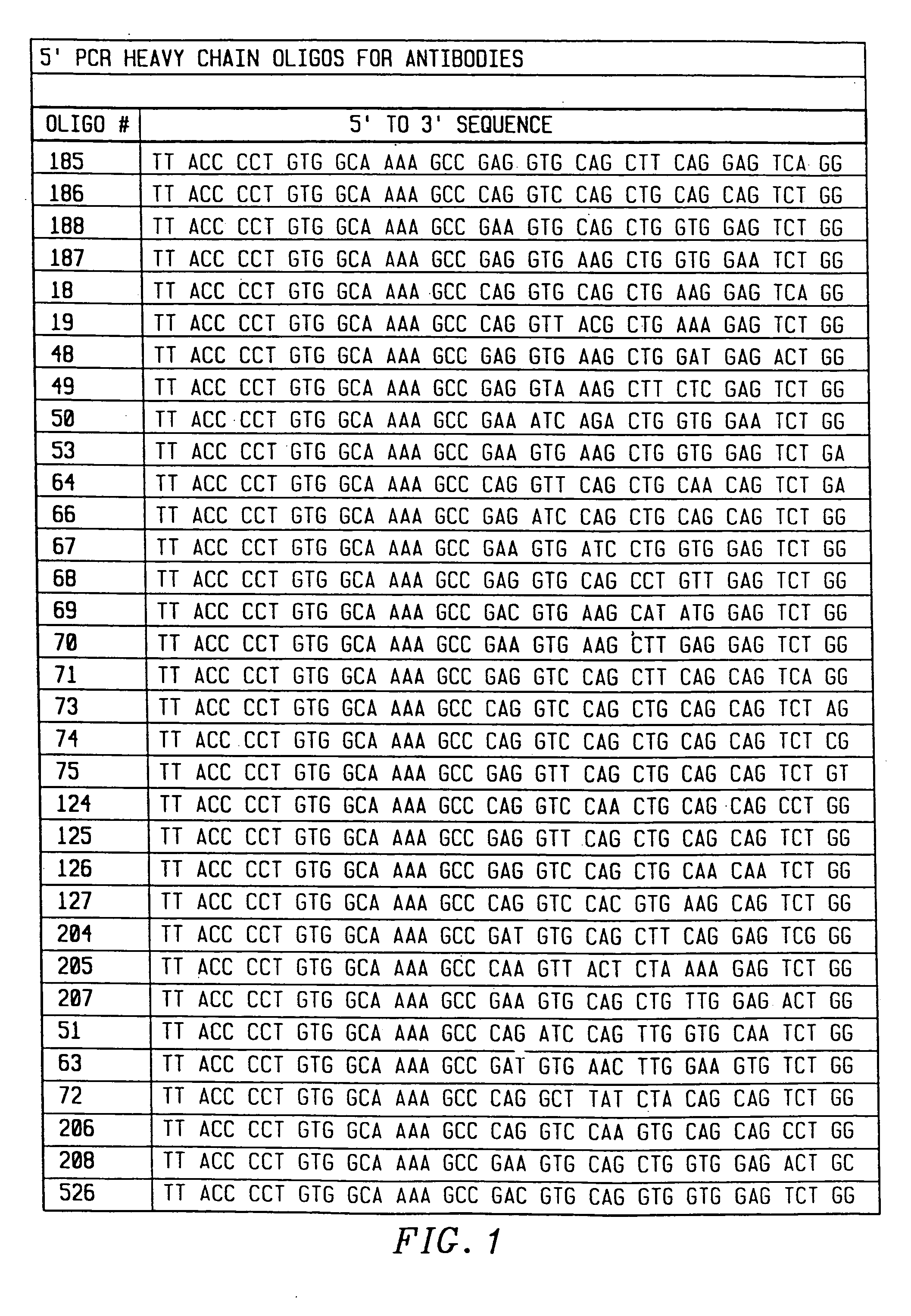
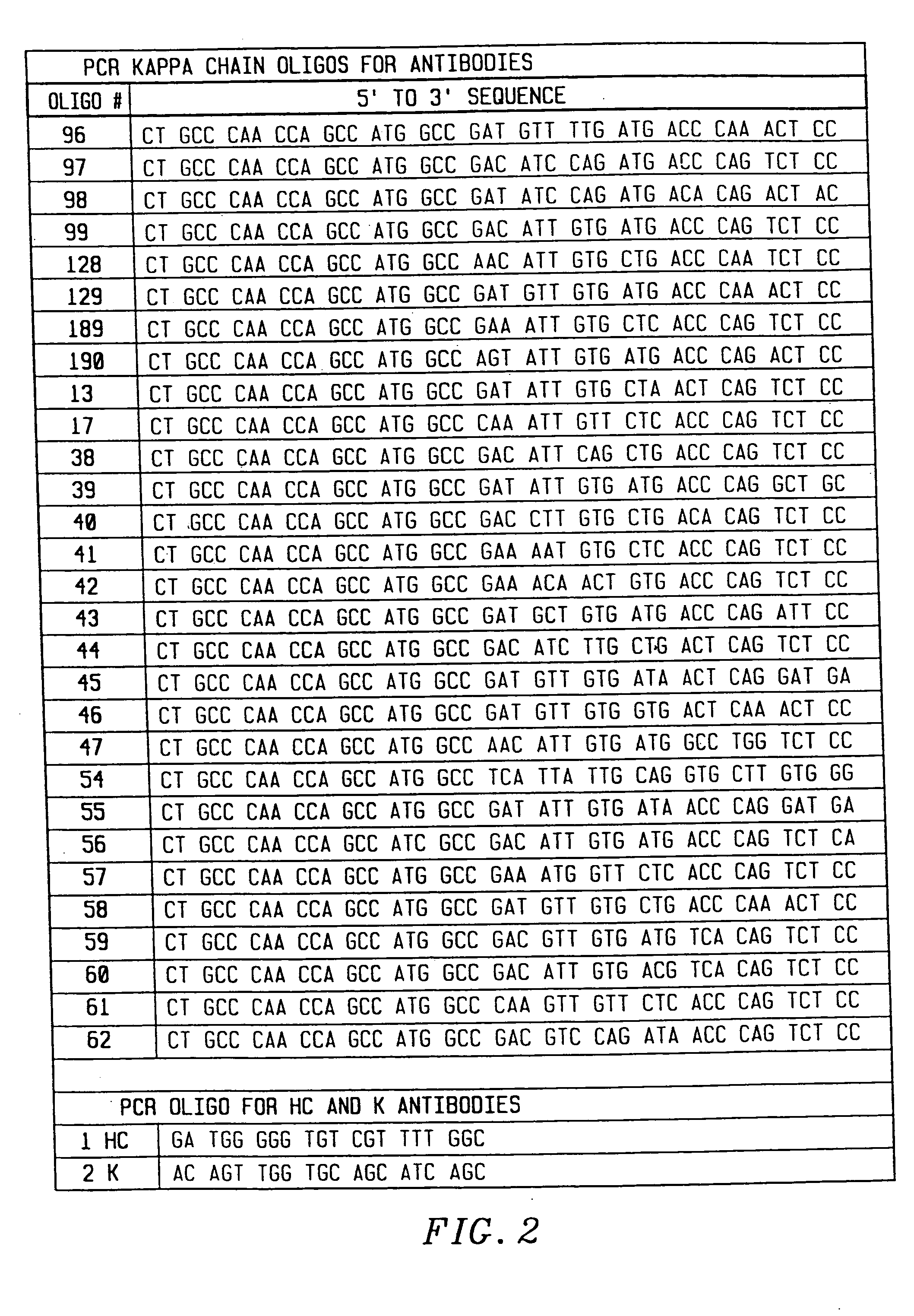
[0149] To amplify substantially all of the H and L chain genes using PCR, primers were chosen that corresponded to substantially all published sequences. Because the nucleotide sequences of the amino terminals of H and L contain considerable diversity, 33 oligonucleotides were synthesized to serve as 5′ primers for the H chains (FIG. 1), and 29 oligonucleotides were synthesized to serve as 5′ primers for the kappa L chains (FIG. 2). The 5′ primers were made according to the following criteria. First, the second and fourth amino acids of the L chain and the second amino acid of the heavy chain were conserved. Mismatches that changed the amino acid sequence of the antibody were allowed in any other position. Second, a 20 nucleotide sequence complementary to the M13 uracil template was synthesized on the 5′ side of each primer. This sequence is different between the H and L chain primers, corresponding to 20 nucleotides on the 3′ side of the pelB signal seq...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Metallic bond | aaaaa | aaaaa |
| Level | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com