Selection of host cells expressing protein at high levels
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Construction of STAR67 Vectors
[0160] A novel anti-repressor (STAR) sequence was isolated using a genetic screen as described in WO 03 / 004704, and this novel sequence was coined STAR67. The effects of STAR67 on expression of transgenes in mammalian cell lines were tested. Here we describe the construction of the various constructs.
Materials and Methods
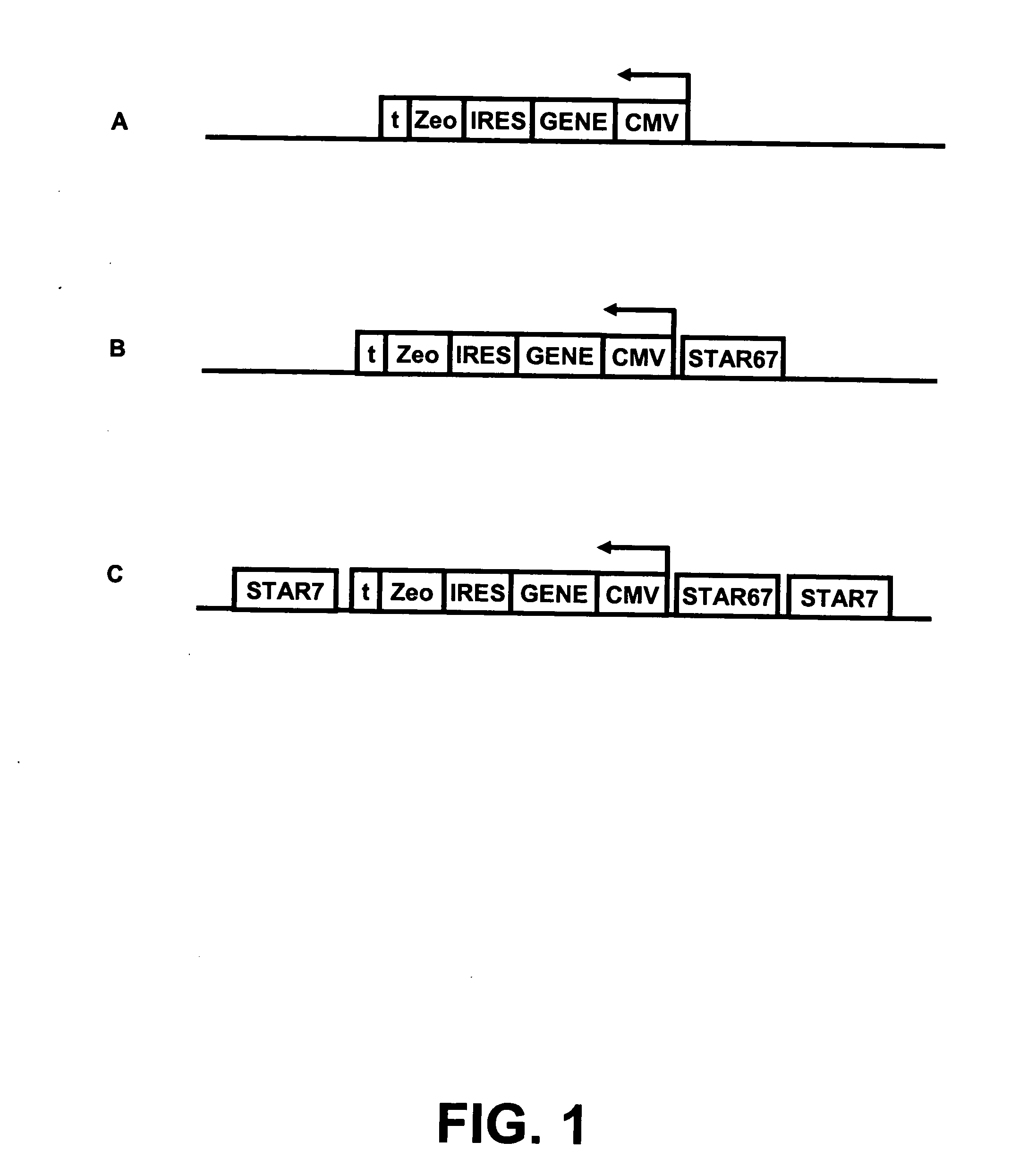
[0161] Three plasmids were created (FIG. 1):
[0162] A) CMV-d2EGFP-ires-Zeo (CMV Control),
[0163] B) STAR67-CMV-d2EGFP-ires-Zeo (CMV-STAR67),
[0164] C) STAR7-STAR67-CMV-d2EGFP-ires-Zeo-STAR7 (CMV-STAR67 7 / 7)
[0165] The construction of construct A is described below. Plasmid pd2EGFP (Clontech 6010-1) was modified by insertion of a linker at the BsiWI site to yield pd2EGFP-link. The linker, made by annealing oligonucleotides GTACGGATATCAGATCTTTAATTAAG (SEQ. ID. NO. 67) and GTACCTTAATTAAAGATCTGATAT (SEQ. ID. NO. 68), introduced sites for the PacI, BglII, and EcoRV restriction endonucleases. This created the multiple cloning site MCSII f...
example 2
STAR67 Enhances the Expression Level from CMV, EF1α and UB6 Promoters in Stably Transfected CHO Cells
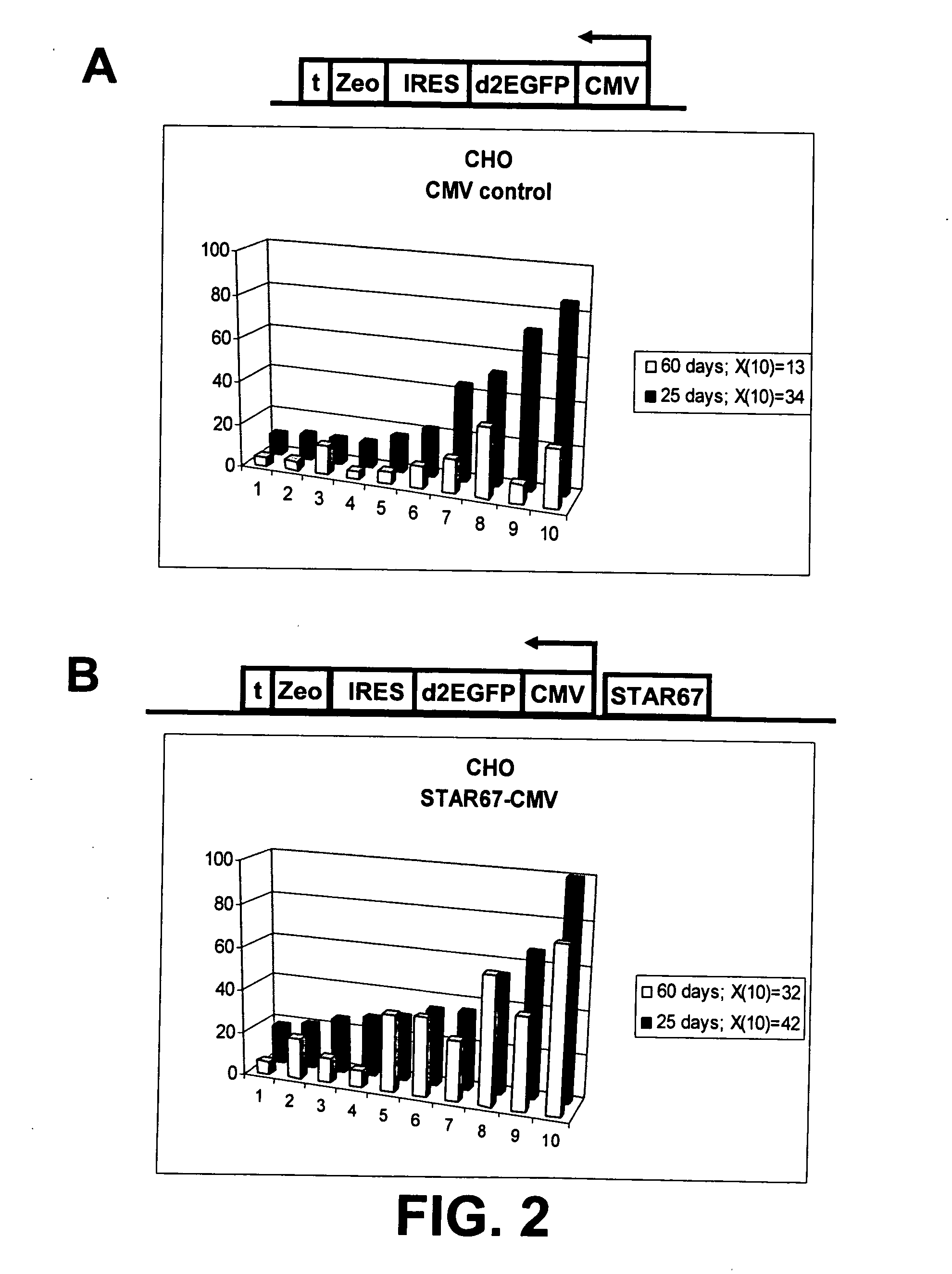
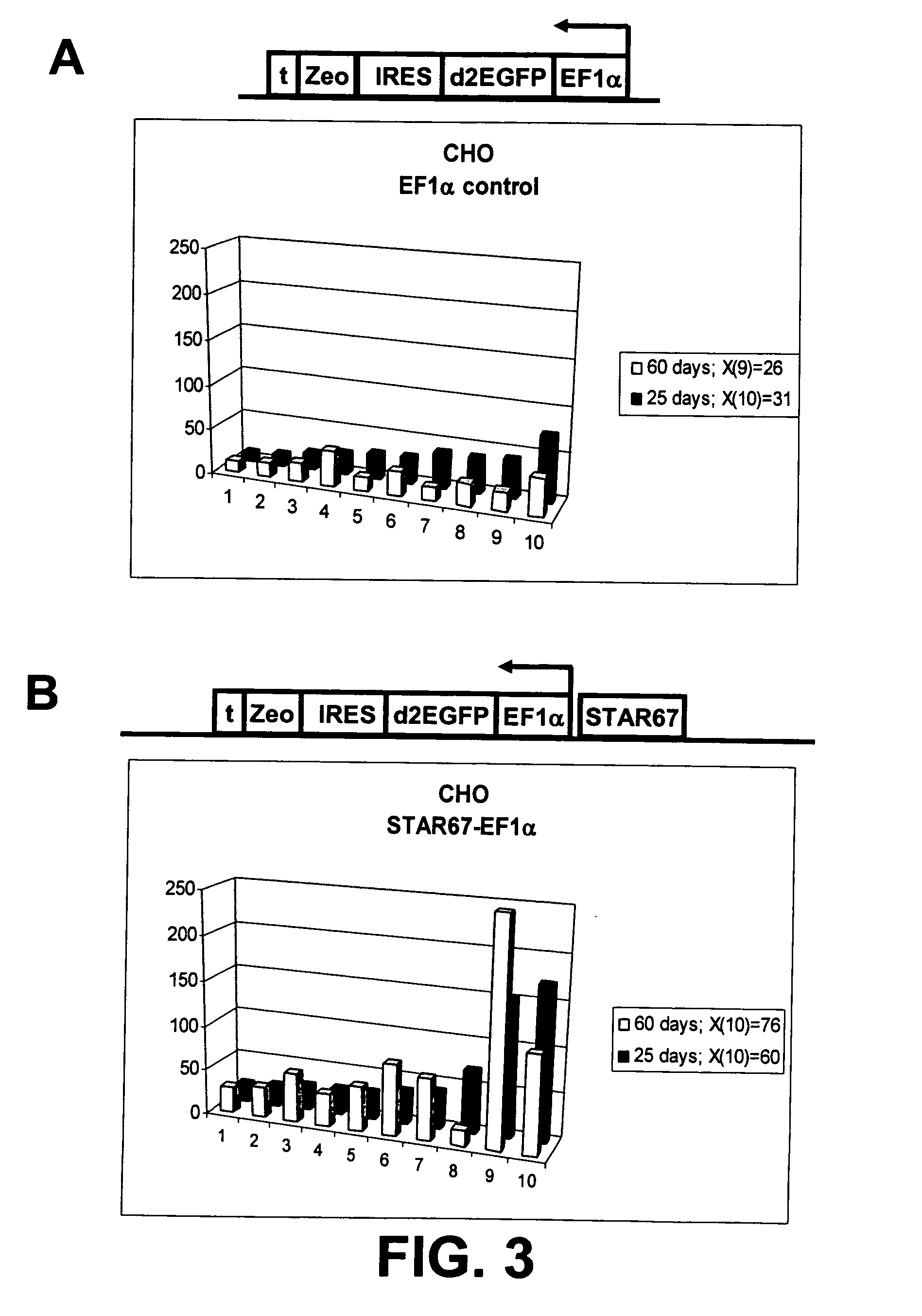
[0169] We tested whether the presence of STAR67 adjacent to the CMV, EF1α and UB6 promoters influences the expression level of these promoters in CHO cells. The constructs A and B (FIG. 1) described in Example 1 are used for this purpose, modified for the respective promoters:
[0170] 1 CMV-d2EGFP-ires-Zeo (CMV Control)
[0171] 2 STAR67-CMV-d2EGFP-ires-Zeo (CMV-STAR67)
[0172] 3 EF1α-d2EGFP-ires-Zeo (EF1α Control)
[0173] 4 STAR67-EF1α-d2EGFP-ires-Zeo (EF1α-STAR67)
[0174] 5 UB6-d2EGFP-ires-Zeo (UB6 Control)
[0175] 6 STAR67-UB6-d2EGFP-ires-Zeo (UB6-STAR67).
Materials and Methods
[0176] The UB6 and EF1α promoters were exchanged for the CMV promoter in the plasmids described in FIG. 1. The UB6 promoter was cloned as follows. A DNA 0.37 kb stuffer from the pd2EGFP plasmid was amplified by PCR, as described in example 1, using primers identified by SEQ. ID. NOs. 69 and 70. The resulting DNA...
example 3
STAR67 Enhances the Expression Level from CMV, EF1α and UB6 Promoters in Stably Transfected PER.C6 Cells
[0183] We tested whether the presence of STAR67 adjacent of the CMV, EF1α and UB6 promoters influences the expression level of these promoters in another cell type than CHO cells, namely human PER.C6 cells. The same constructs as in example 1 were used.
Materials and Methods
Transfection, Culturing and Analysis of PER.C6 Cells
[0184] PER.C6® cells were cultured in DMEM medium+pyridoxine+9% Foetal Bovine Serum (Non-Heat Inactivated), 8.9 mM MgCl2 100 U / ml penicillin, and 100 micrograms / ml streptomycin at 37° C. / 10% CO2. Cells were transfected with the plasmids using Lipofectamine 2000 (Invitrogen) as described by the manufacturer. Briefly, cells were seeded to 6-wells and grown overnight to 70-90% confluence. Lipofectamine reagent was combined with plasmid DNA at a ratio of 15 microliters per 3 microgram (e.g. for a 10 cm Petri dish, 20 micrograms DNA and 120 microliters Lipofec...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Electrical resistance | aaaaa | aaaaa |
| Antimicrobial properties | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com