Nucleic acid ligands to integrins
a technology of integrins and ligands, applied in the field of integrin proteins, can solve the problems of limiting the growth and metastasis of cancerous lesions, solid tumors cannot grow to significant size without an independent blood supply, and the affinity of integrins for fibrinogen is very low on these cells
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Isolation of Integrins and Integrin Ligands
[0070]αvβ3 integrin was isolated from human placenta and purified by immunoaffinity chromatography essentially as described by (Smith and Cheresh (1988) J. Biol. Chem. 263:18726-31). In brief, human placentas were diced and the tissue fragments were extracted in a buffer containing 100 mM octyl-β-D-glucopyranoside detergent (Calbiochem, San Diego, Calif.). The extract was cleared by centrifugation and applied to an immunoaffinity column αvβ3-specific monoclonal antibody LM609 affixed to Affi-Gel 10, (Chemicon International, Inc., Temecula, Calif.)). Protein bound to the column was eluted with a low-pH buffer and fractions were immediately neutralized and analyzed for integrin content by SDS-polyacrylamide gel electrophoresis. Integrin-containing fractions were pooled and aliquots of the purified material were stored at −80° C. Purified human αvβ3 was also purchased from Chemicon International, Inc, as was human αvβ5 integrin. αIIbβ3 and fi...
example 2
Generating Nucleic Acid Ligands to Integrins Using the SELEX Method
[0071] A DNA template library of sequence:
5′-ttatacgactcactatagggagacaagaataaac(SEQ ID NO:1)gctcaannnnnnnnnnnnnnnnnnnnnnnnnnnnnnnnnnnnnnnnttcgacaggaggctcacaacaggc-3′
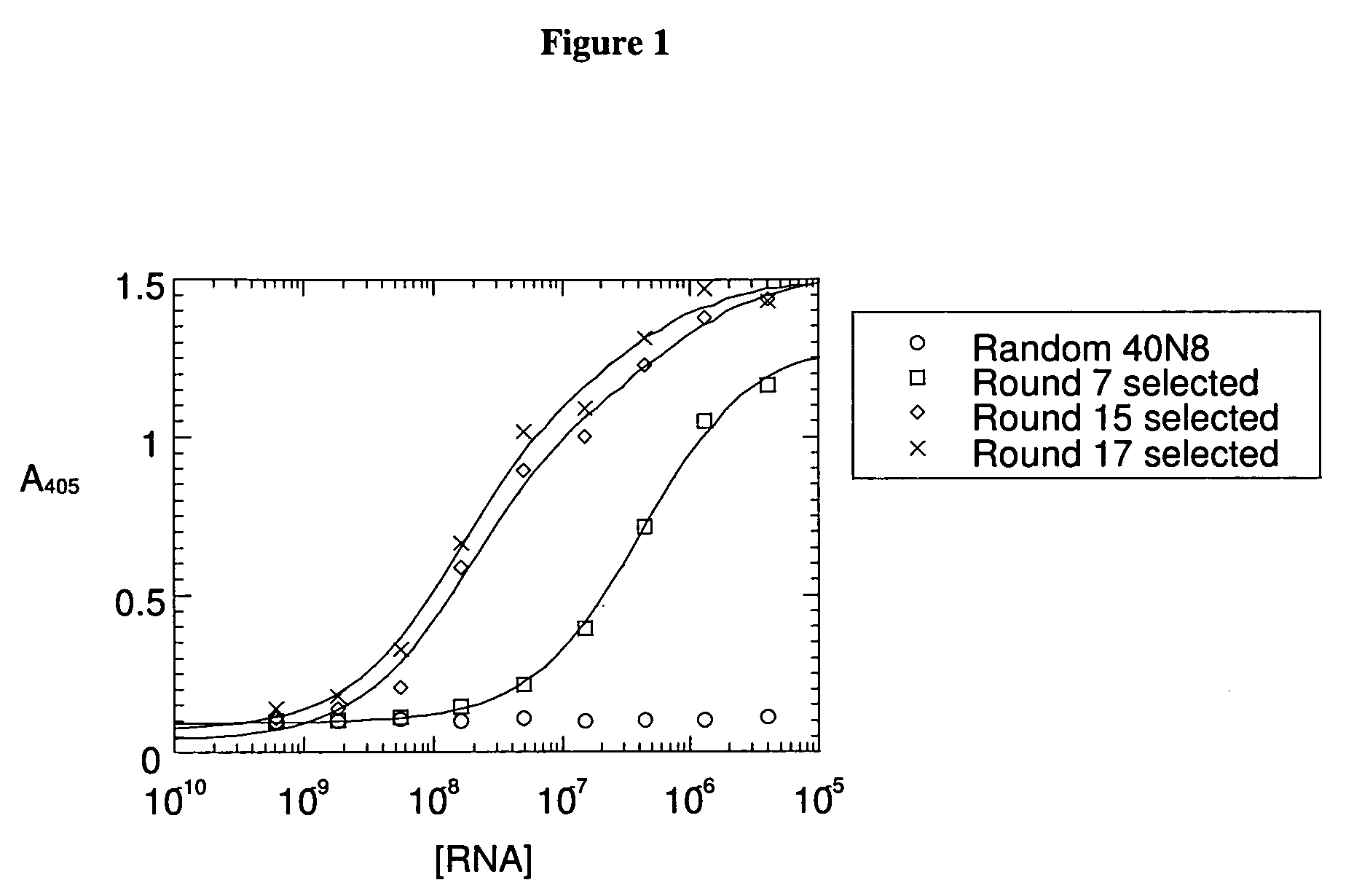
was prepared by chemical synthesis. The italicized nucleotides correspond to a T7 RNA polymerase promoter. There are 40 n residues (a,g,t, or c). A short DNA primer “3N8”:5′-gcctgttgtgagcctcctgtcgaa-3′ (SEQ ID NO:2) was annealed to the template and extended using Klenow DNA polymerase (New England Biolabs, Beverly, Mass.). The double-stranded DNA product served as a product for T7 RNA polymerase transcription (enzyme obtained from Enzyco, Inc., Denver, Colo.) to generate a library of random-sequence RNAs. 2′-fluoro-CTP and -UTP were used in place of the 2′-OH-pyrimidines.
[0072] For application of the SELEX process to αvβ3 integrin, the purified protein was diluted 1000-fold from detergent-containing storage buffer into 50 mM MES (2-[N-morpholino]etha...
example 3
Specificity of the Nucleic Acid Ligands to Integrins
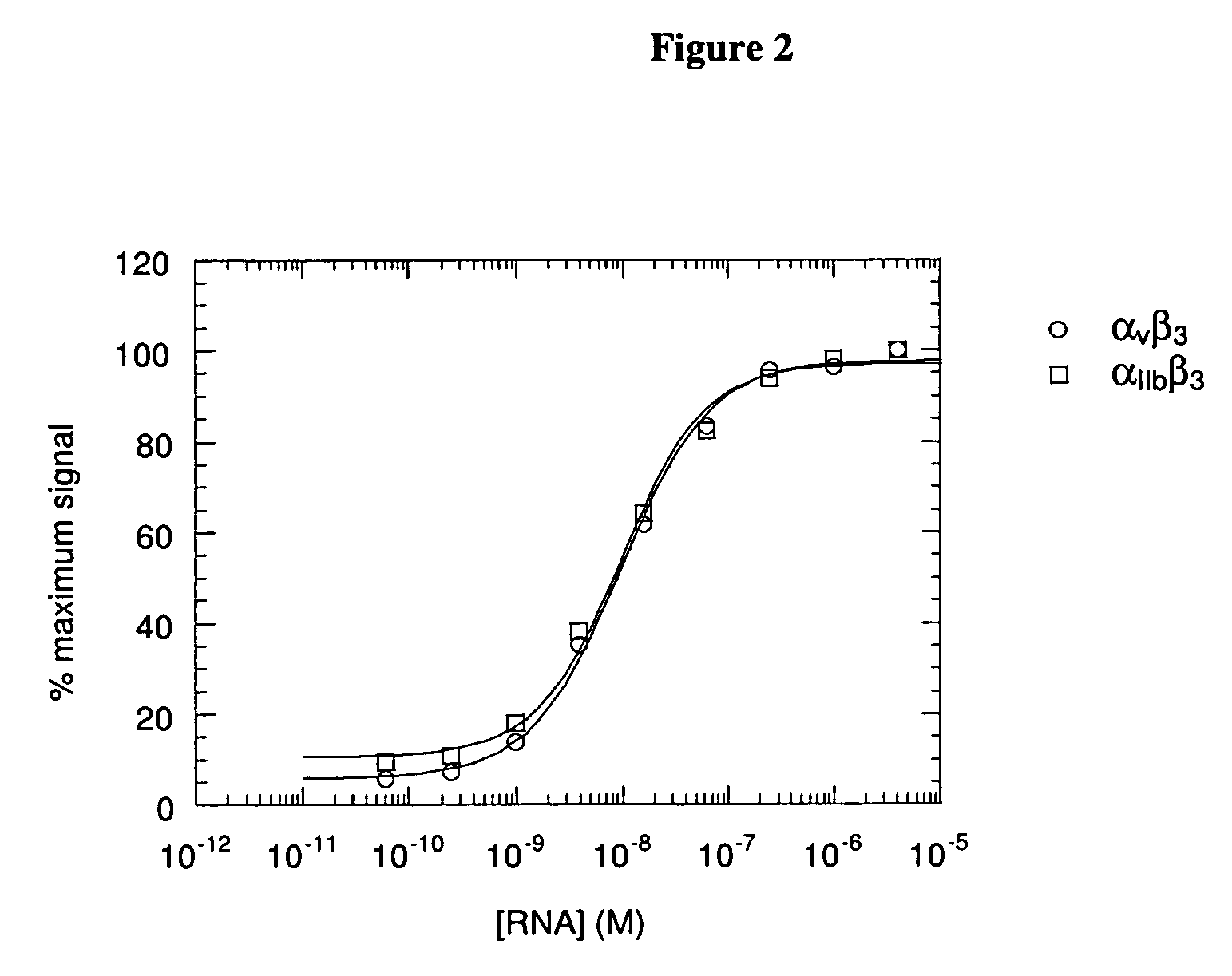
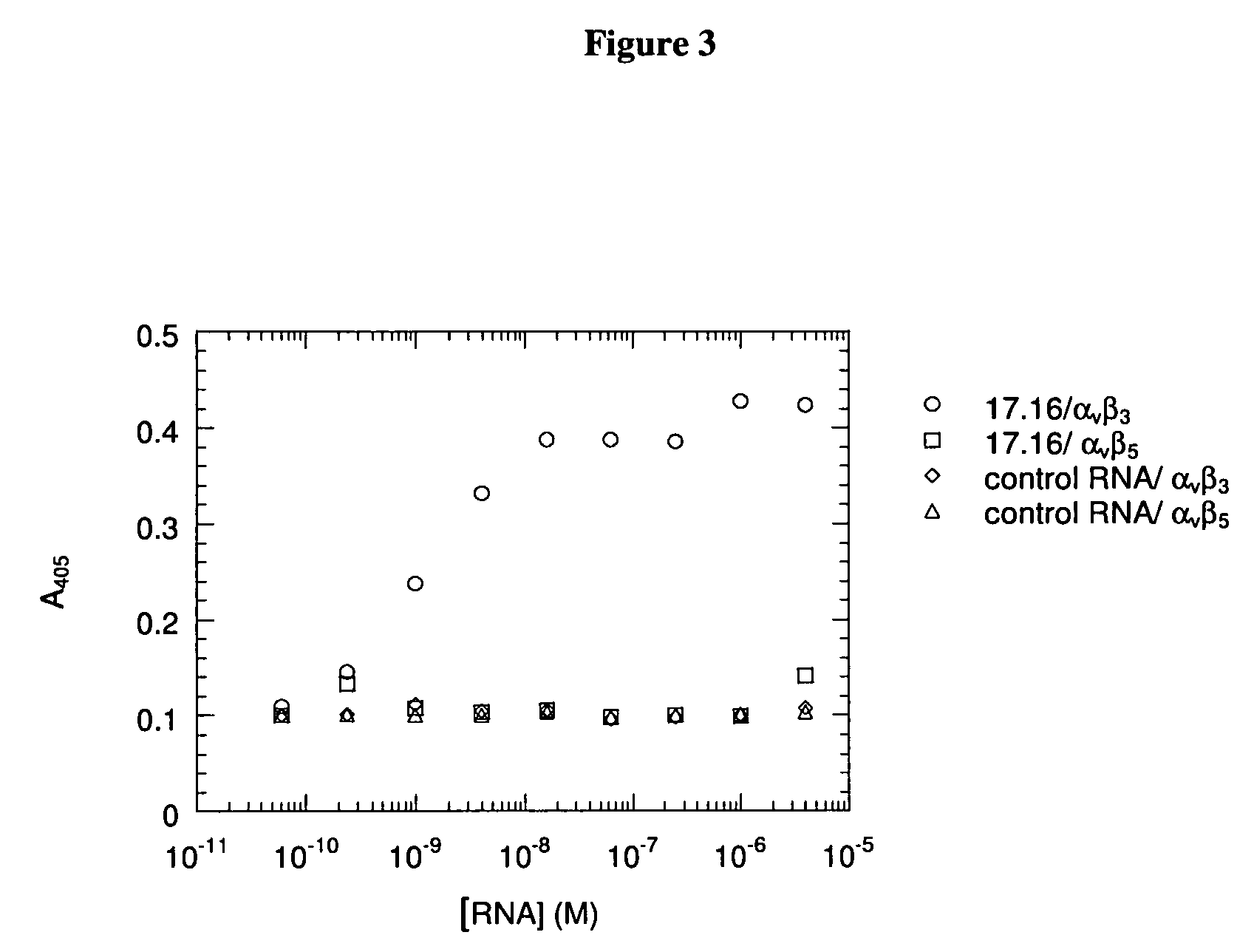
[0077] In general, aptamers selected for high-affinity binding to a particular target protein show relatively weak binding to other related proteins, except in cases where the degree of homology is very high (for example, see (Green et al. (1996) Biochem. 35:14413-24; Ruckman et al. (1998) J. Biol. Chem. 273:20556-67)). Significant homology exists within the families of integrin alpha and beta sub-units, and both alpha and beta sub-units are shared among members of the integrin superfamily. Thus, it was of interest to assess the relative affinity of the αvβ3 aptamers for closely related integrins. The affinities were determined using the methods described above. The Family 1 aptamer 17.16 (SEQ ID NO:60) was chosen as a representative of the major sequence family. FIG. 2 shows that aptamer 17.16 bound with identical affinity to purified, utilized αvβ3 and to the platelet integrin, αIIbβ3 in a 96-well plate binding assay. Although t...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molar density | aaaaa | aaaaa |
| Molar density | aaaaa | aaaaa |
| Molar density | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com