Systems for gene targeting and producing stable genomic transgene insertions
a technology of gene targeting and stable insertion, applied in the field of gene targeting and producing stable genomic transgene insertion, to achieve the effect of facilitating the stabilization process and enhancing the efficiency of cassette exchang
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
embodiment 1
Excision-Competent Stabilization Vectors
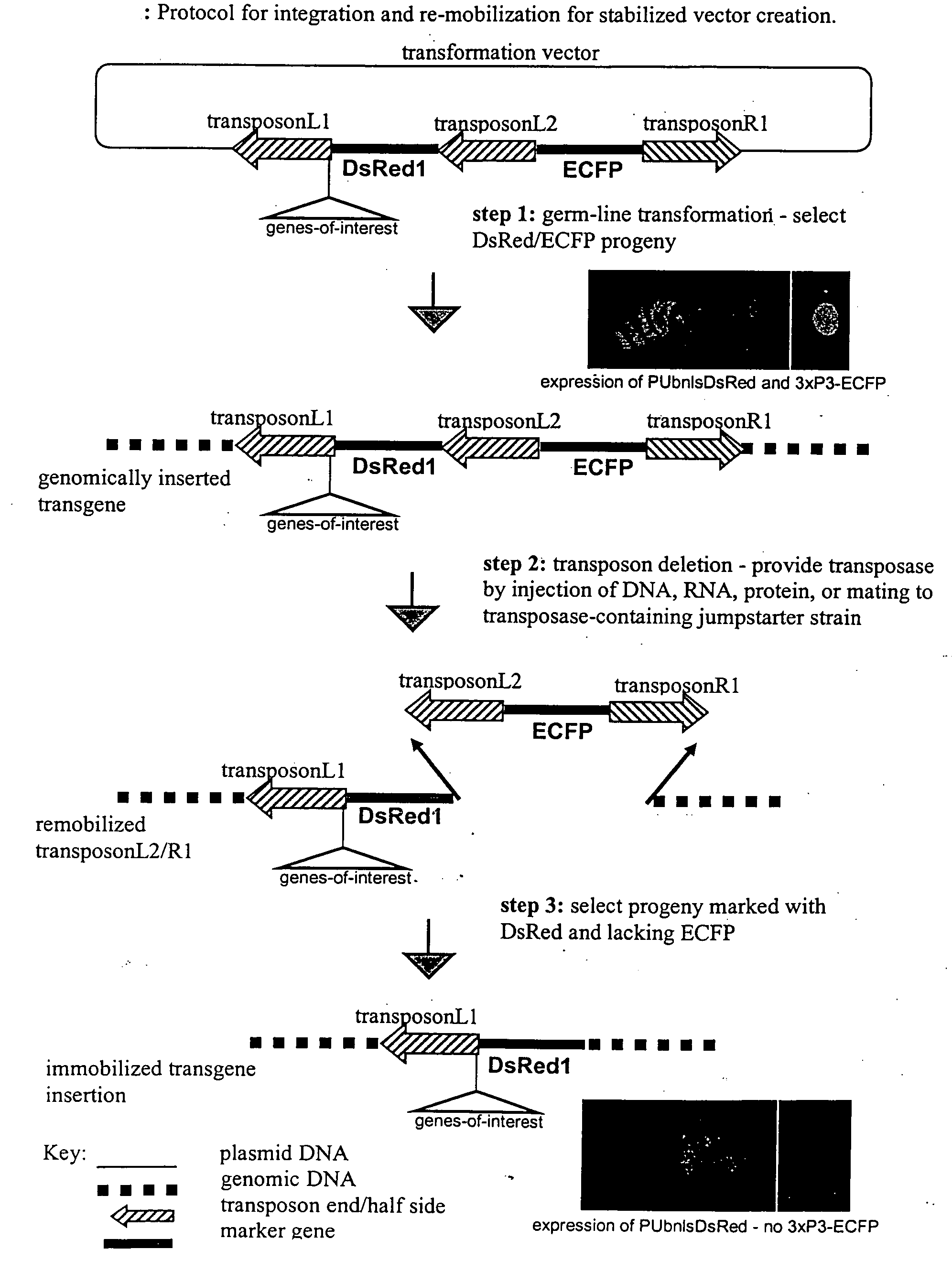
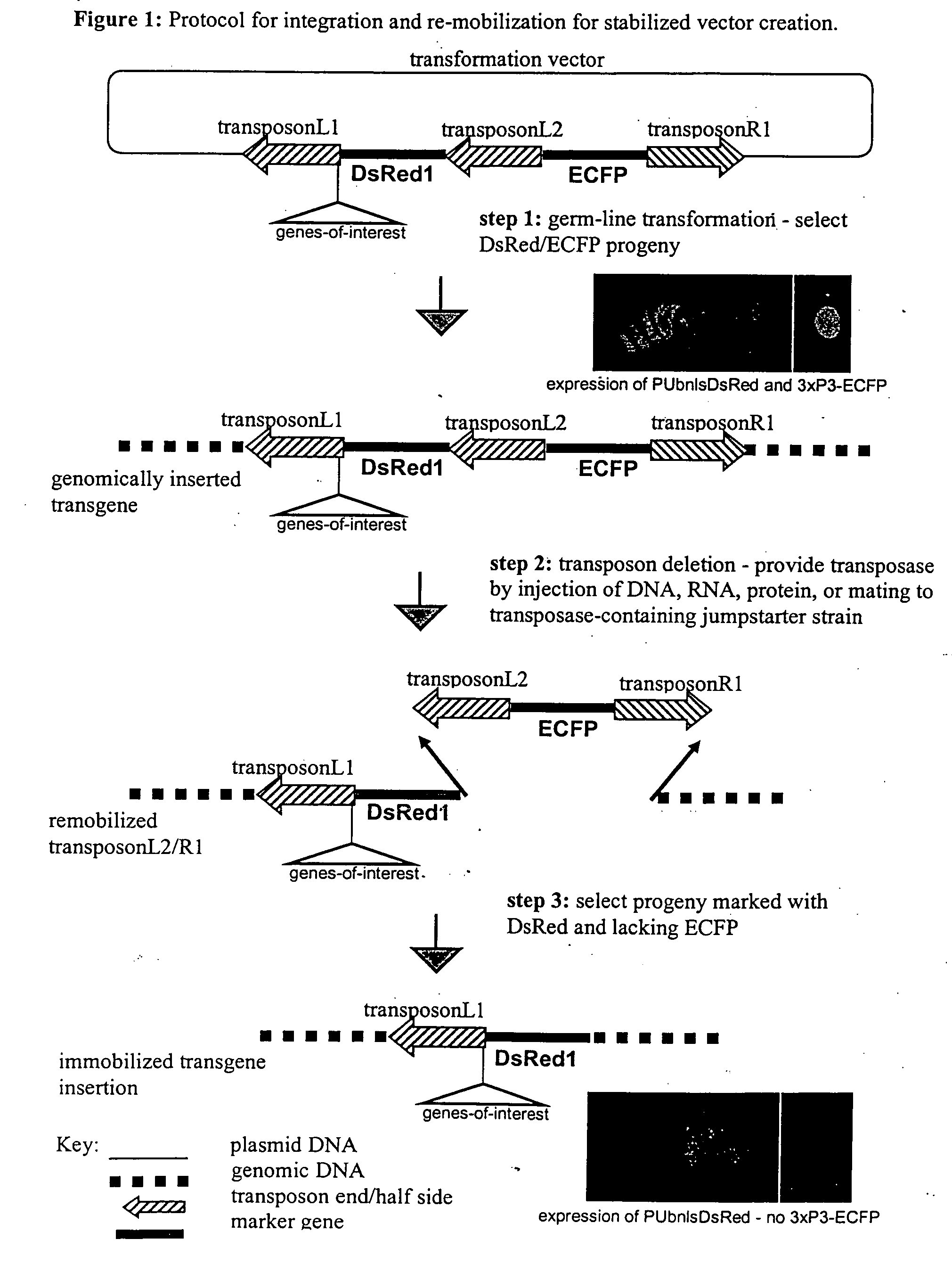
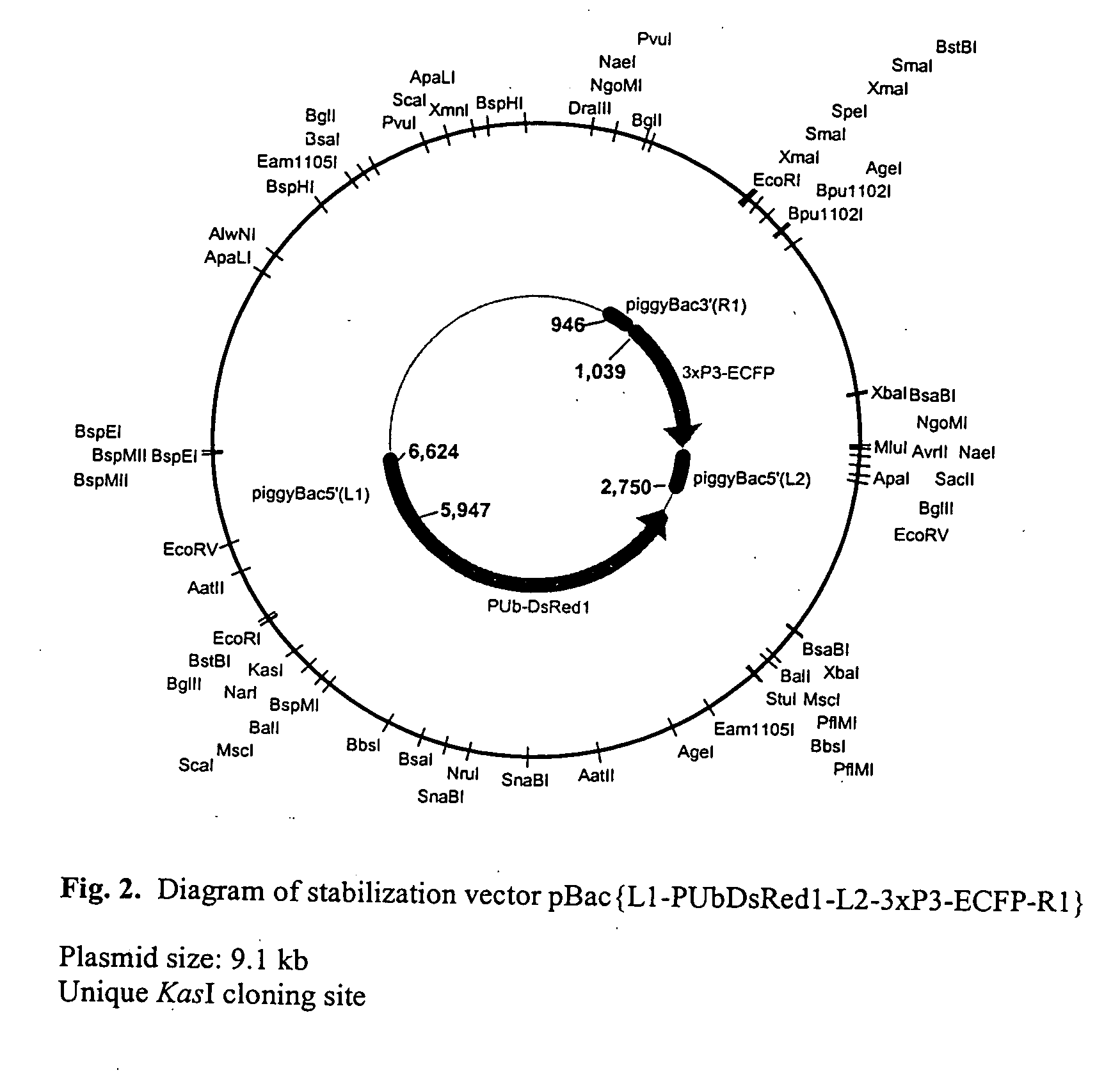
[0029]The experimental steps for the method are described in FIG. 1, and the structure of the excision competent transformation vector, pBac{L1-PUbDsRed1-L2-3×P3-ECFP-R1 }, is described in FIG. 2. Integration and re-mobilization of the vector was verified by PCR and sequence analysis described in FIG. 3. pBac{L1-PUbDsRed1-L2-3×P3-ECFP-R1} was constructed based on the transposable element “piggyBac” (see U.S. Pat. No. 6,218,185, the contents of which are incorporated herein by reference). Conventional piggyBac-based transformation vectors (see WO 01 / 14537 and WO 01 / 12667, the contents of which are incorporated herein by reference) typically contain piggyBac-half sides or parts thereof, including 5′ piggyBac terminal sequences (referred to as piggyBacL) and 3′ piggyBac terminal sequences (referred to as piggyBacR), which flank a transformation marker gene and a cloning site to insert the genes-of-interest. (see Handler, A. M., 2001. A current pe...
embodiment 2
Conditional Excision-Competent Transformation Vectors
[0036]The structure of the conditional excision-competent transformation vector, pBac_STBL, as well as the experimental steps are depicted schematically in FIGS. 4 and 6. pBac_STBL is based on the transposable element “piggybac” (see U.S. Pat. No. 6,218,185, the contents of which are incorporated herein by reference) and is a modified version of pBac{L1-PUbDsRed1-L2-3×P3-ECFP-R1 }. In pBac-STBL the internal transposon half-side (R2) is a duplication of the piggyBac 3′-end, and it is in reverse, or opposite, orientation to R1. In addition, it is flanked in upstream and downstream positions by FRT (FLP recombinase target) sites in opposite directions that create an inversion by recombination in the presence of FLP recombinase (see FIGS. 4 and 6). Therefore, in this vector, only the piggyBacL1 and R1 half sides and intervening DNA can integrate, but re-mobilization of piggyBacR2 together with piggyBacL1 or piggyBacR1 should not be po...
embodiment 3
RMCE with Subsequent Transposon Deletion
[0054]The RMCE-acceptor plasmid, pBac{3×P3-FRT-ECFP-linotte-FRT3} (FIG. 8), is a piggyBac-based transformation vector that was provided additionally with a DNA exchange cassette. This cassette consists of two heterospecific FRT sites (referred to as FRT and FRT3 equivalent to F and F3 (published in European Patent No. EP 0 939 120 A1, the contents of which are incorporated herein by reference)) in parallel orientation.
[0055]European Patent No. EP 0 939 120 A1 (see page 2, line 50 to page 3, line 6) teaches the technology of the RMCE reaction:[0056]“Recombinases such as FLP and Cre have emerged as powerful tools to manipulate the eucaryotic genome (Kilby, N. J., Snaith, M. R., Murray, J. A. H. (1993). Site-specific recombinases: tools for genome engineering. Trends Genet. 9, 413-421, and Sauer B. (1994). Site-specific recombination: developments and applications. Curr. Opin. Biotechnol. 5, 521-527, the contents of which are incorporated by refe...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
| fluorescent | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com