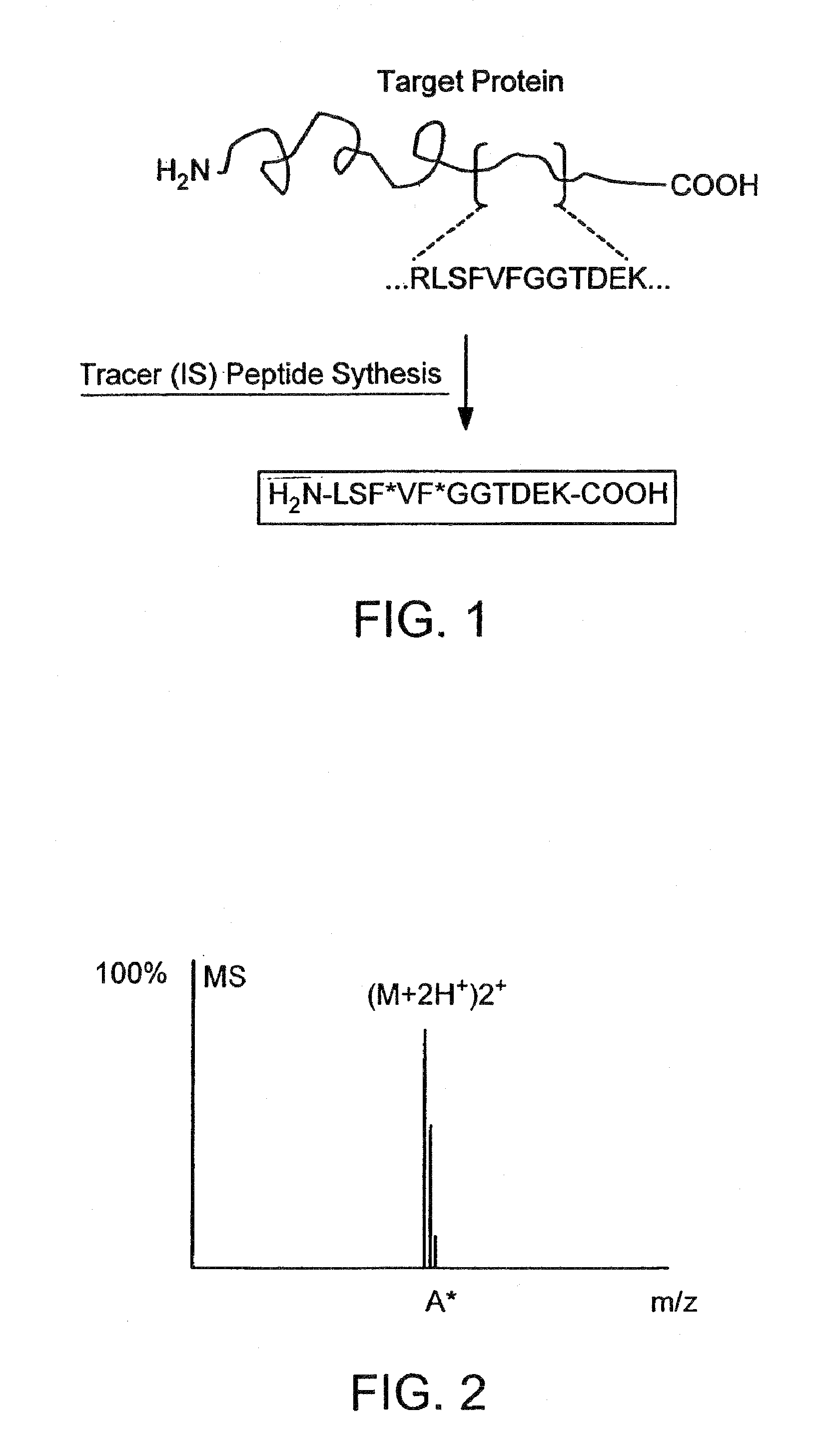
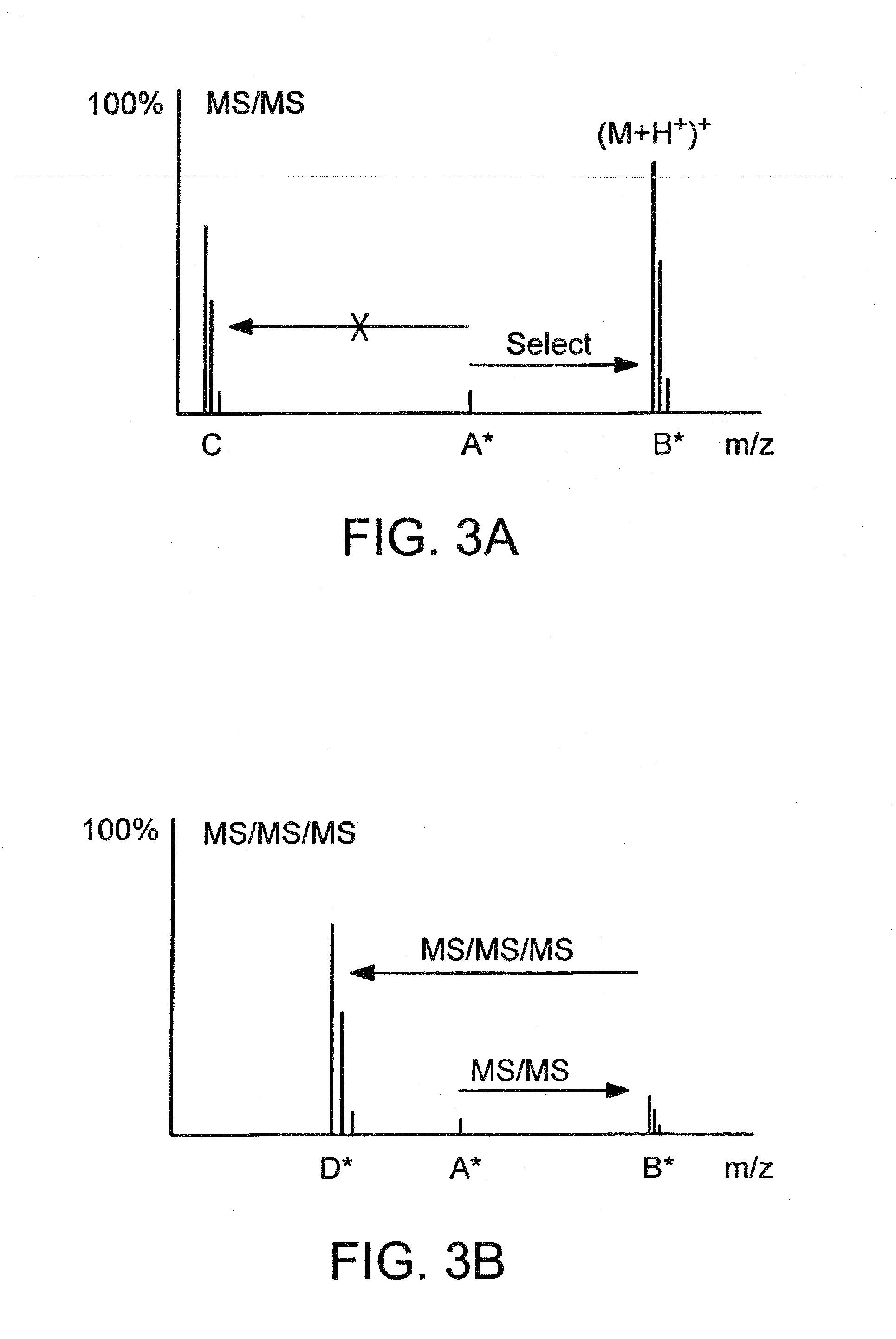
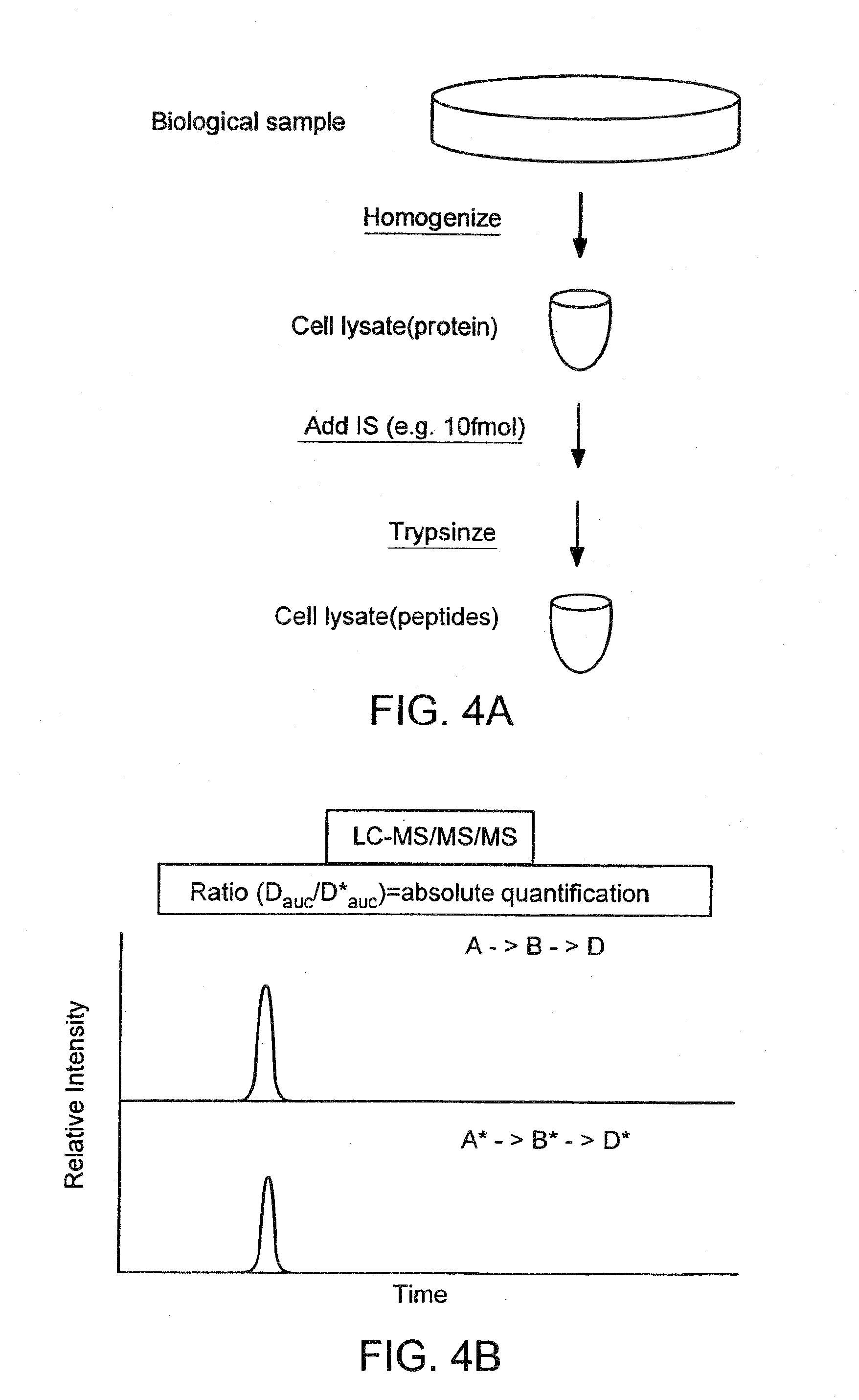
Detection and quantification of modified proteins
a technology of modified proteins and quantification methods, applied in the field of detection and quantification of modified proteins, can solve the problems of difficult to easily determine the quantity of analyzed proteins, severely limit cross-reactivity, and substantially alleviate the dynamic range problem, and achieve the effect of rapid and high-throughput analysis of proteomes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Preparation of Ubiquitin-Conjugates from S. cerevisiae
[0220]Isolation and identification of yeast ubiquitin-conjugates was accomplished as illustrated in FIG. 8. 100 mg of whole yeast lysates were harvested from cells growing through log phase (OD610 1-1.5) from two strains of yeast differing in the expression of 6×His-tagged ubiquitin. Strain SUB592 (JSY171), expressing tagged ubiquitin, and control strain, SUB280 (Spence, et al., 2000, Cell 102: 67-76), were grown to log phase and lysed in buffer A (10 mM Tris, pH 8.0, 0.2 M NaH2PO4, 8M Urea) using glass beads. A Ni2+-NTA-agarose column (Qiagen, Chatsworth, Calif.) was loaded with the clarified lysates, sequentially washed with 30 volumes (bed volume) of buffer A twice, 3 volumes of buffer B (10 mM Tris, pH 6.3, 0.1 M NaH2PO4, 8M Urea), and eluted with 3 volumes of buffer C (10 mM Tris, pH 4.5, 0.1 M NaH2PO4, 8M Urea). A portion (0.5%) of eluted polypeptides was examined by SDS-PAGE and silver staining. The remaining polypeptides...
PUM
| Property | Measurement | Unit |
|---|---|---|
| mass | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
| mass difference | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com