Methods for quantitative genetic analysis of cell free DNA
a cell-free dna and quantitative technology, applied in the field of cell-free dna quantitative genetic analysis, can solve the problems of lack of reliable and robust molecular analysis methods in the analysis of genetic diseases, dna sequencing as a molecular diagnostic tool has been generally limited to the coding exons of one or two genes, and requires direct access to tumor tissues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Accurate Detection of Rare Mutations using Targeted Sequence Capture Technology
Purpose
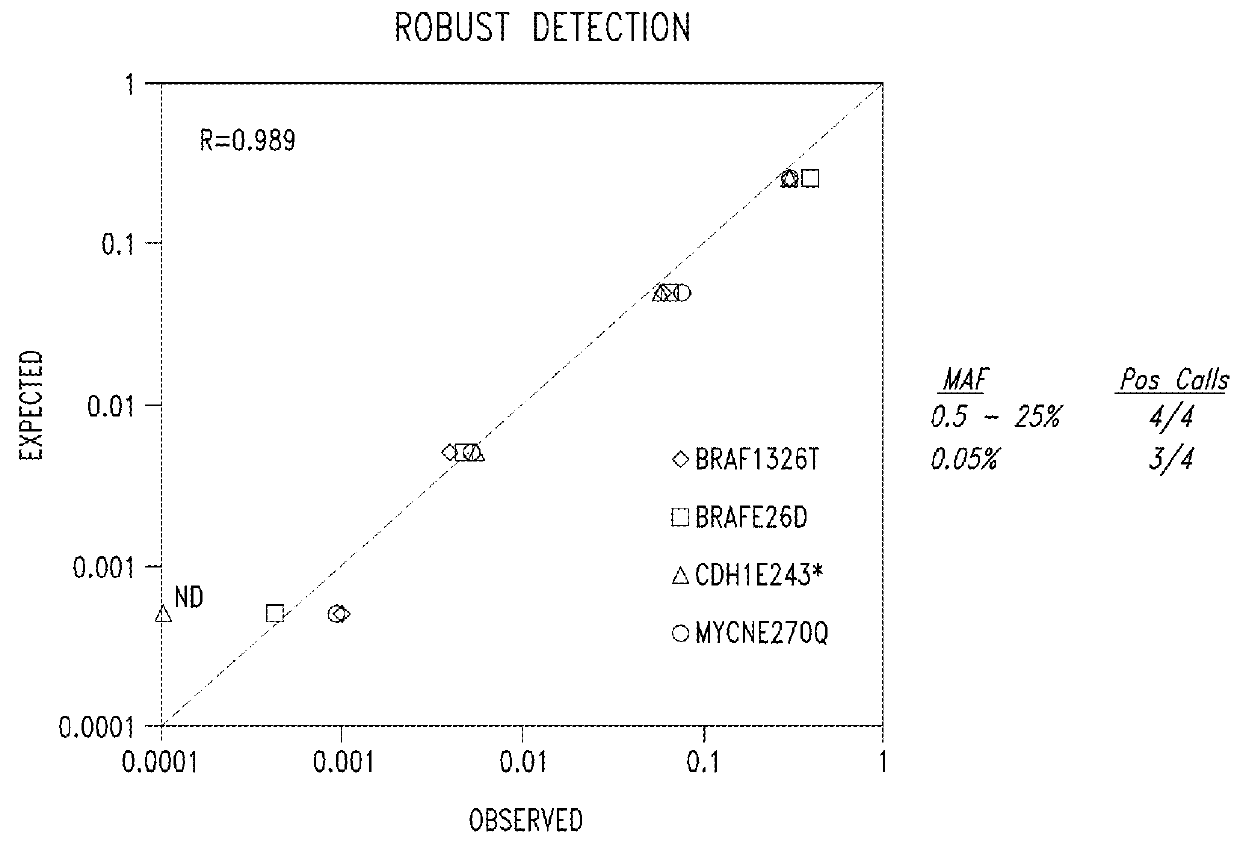
[0221]The purpose of this experiment was to provide a direct proof-of-principle demonstration of rare variant detection using targeted sequence capture technology.
Background
[0222]Target sequence capture technology provides quantitative, sequence-based genetic analysis of nucleic acids and can be exploited to perform a combined mutational and copy number analysis of drug metabolism genes. The present inventors used targeted sequence capture technology and subsequence genetic analysis to detect rare sequence variants.
[0223]Genomic DNA inputs play a central role in rare variant detection, but quantitative analysis and control of genomic inputs places bounds on the estimated sensitivity of rare variant analysis. A genomic qPCR assay was used by the present inventors to estimate genomic inputs.
[0224]One experimental goal for rare variant analysis is to introduce 10-fold more genomic input as the target ...
example 2
A Novel Probe Design Effective for Comprehensive Sequencing of Target Regions in Highly Fragmented gDNA
Purpose
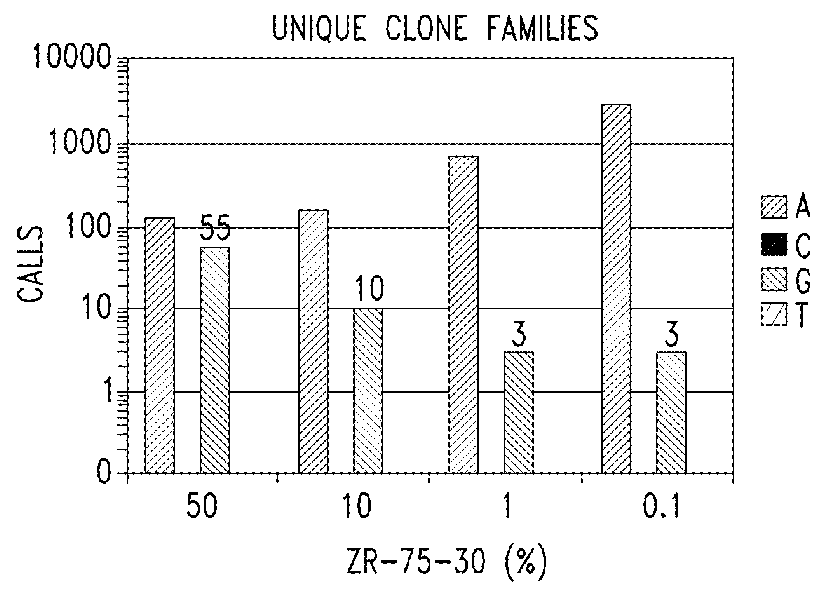
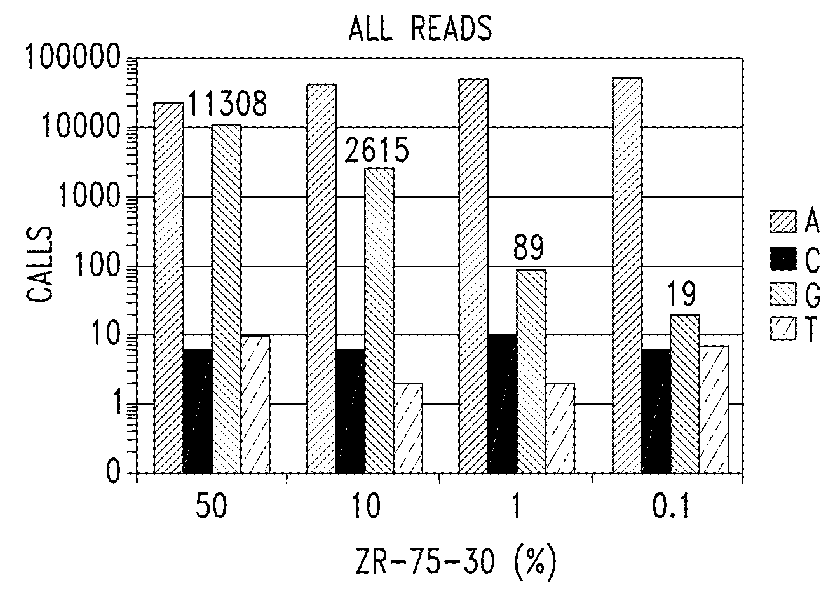
[0234]The purpose of these experiments is to develop an assay system to reliably and reproducibly interrogate circulating DNAs.
Background
[0235]Analysis of circulating DNA from body fluids represents an exciting, but as yet, unrealized opportunity in molecular diagnostics. Genomic DNA is highly intact. Literature suggest that the average size of circulating DNA is about 150 bp, which correlates well to the size of DNAs wrapped around a single nucleosomal histone complex.
Summary
[0236]The technical parameters of targeted sequence capture technology contemplated herein were designed to accommodate highly fragmented DNA and to retain the ability to generate comprehensive sequence coverage of targeted DNA. Capture probe density was increased and the length of capture probe sequences was reduced from 60 nucleotide to 40 nucleotide to minimize uninformative sequence generation in th...
example 3
Genetic Analysis of Circulating DNA
Purpose
[0272]The purpose of this example was to benchmark the genetic analysis of cfDNA using an efficient cloning procedure for cfDNA and target retrieval system.
Background
[0273]While there is tremendous enthusiasm in the scientific and health-care community for “liquid biopsies”—analysis of circulating DNA (cfDNA) for markers associated with potential disease states, there is remarkably little practical information about this potential analyte.
Summary
[0274]Plasma samples collected from healthy donors and individuals suffering from either ovarian or colon cancers were used to perform the genetic analysis of circulating DNA. The amount and the overall character of circulating cfDNA can vary widely from individual to individual. Surprisingly, the present inventors found that cfDNA is readily clonable with an efficiency indistinguishable from highly purified and fragmented genomic DNA; that the fragment size was remarkably consistent, with an average...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperatures | aaaaa | aaaaa |
| Tm | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com