Apparatus and methods for kinetic analysis and determination of nucleic acid sequences
a technology of nucleic acid sequence and apparatus, applied in the field of detection and characterization of nucleic acids, can solve the problem of prohibitively expensive wide-scale deployment across large populations of individuals
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
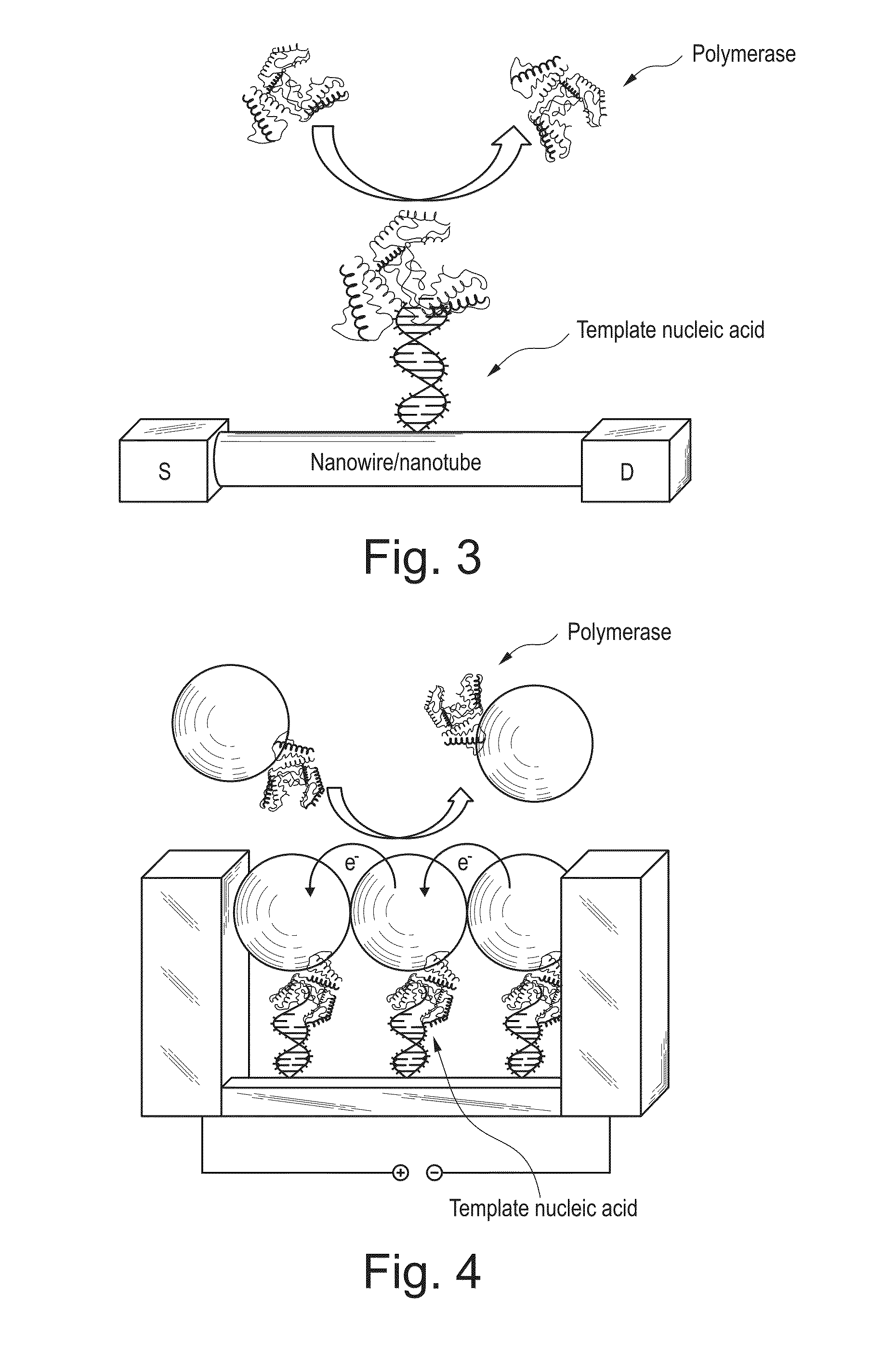
Image
Examples
example i
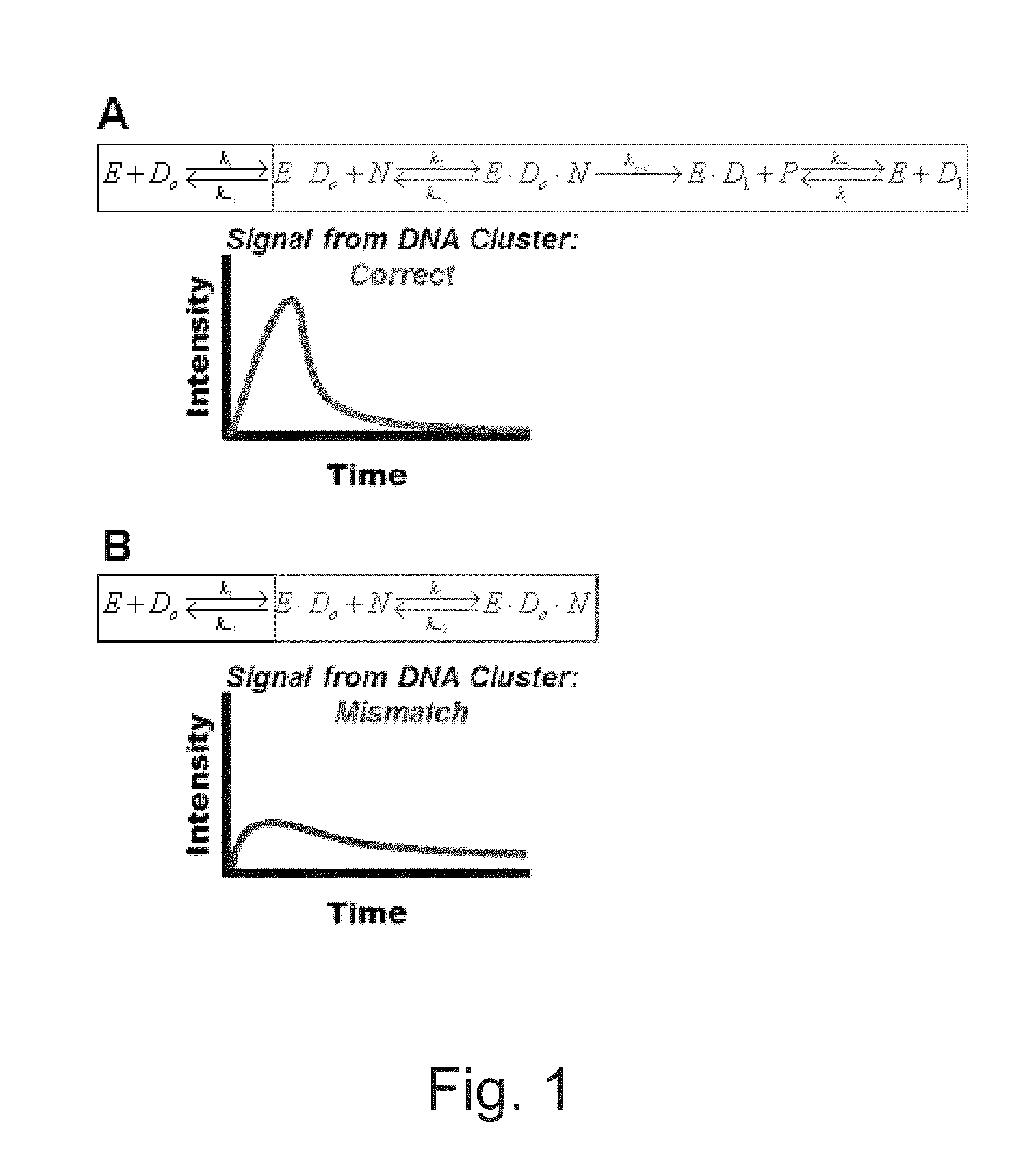
Single Molecule Pattern Sequencing Using Pre-Equilibrium Kinetics Scheme 1
[0139]Glass coverslips were treated with PEG / PEG-biotin according to single molecule protocols as described by Roy, et al., Nature Methods, 5, 507-516 (2008). Coverslips were incubated with 200 pM streptavidin for 15 minutes, washed with 1 ml wash buffer (50 mM Tris pH=7.5; 50 mM NaCl), and incubated for 15 minutes with 200 pM biotinylated DNA template. The target DNA template sequence (SEQ ID NO: 1) is as follows:
Biotin-3′ CTTGCGTGGACACGTTCGCGAACGTGTCCACGCAAGGAATTCG-5′
Surfaces were washed after 15 minutes with 1 ml wash buffer and imaged.
[0140]Cy3-Klenow was prepared as follows. Purified wild-type Klenow exo− was first treated with 10 mM dithiothreitol (DTT) for 30 min at room temperature to reduce disulfide bonds. Next, wild-type Klenow exo− was purified by fractionating with a Sephadex-25 column using the following buffer: 50 mM ACES pH 7.0, 1 M NaCl, 1 mM EDTA, and 0.01% w / v Tween-20. The fractions with wi...
example ii
Single Molecule Pattern Sequencing Using Pre-Equilibrium Kinetics Scheme 2
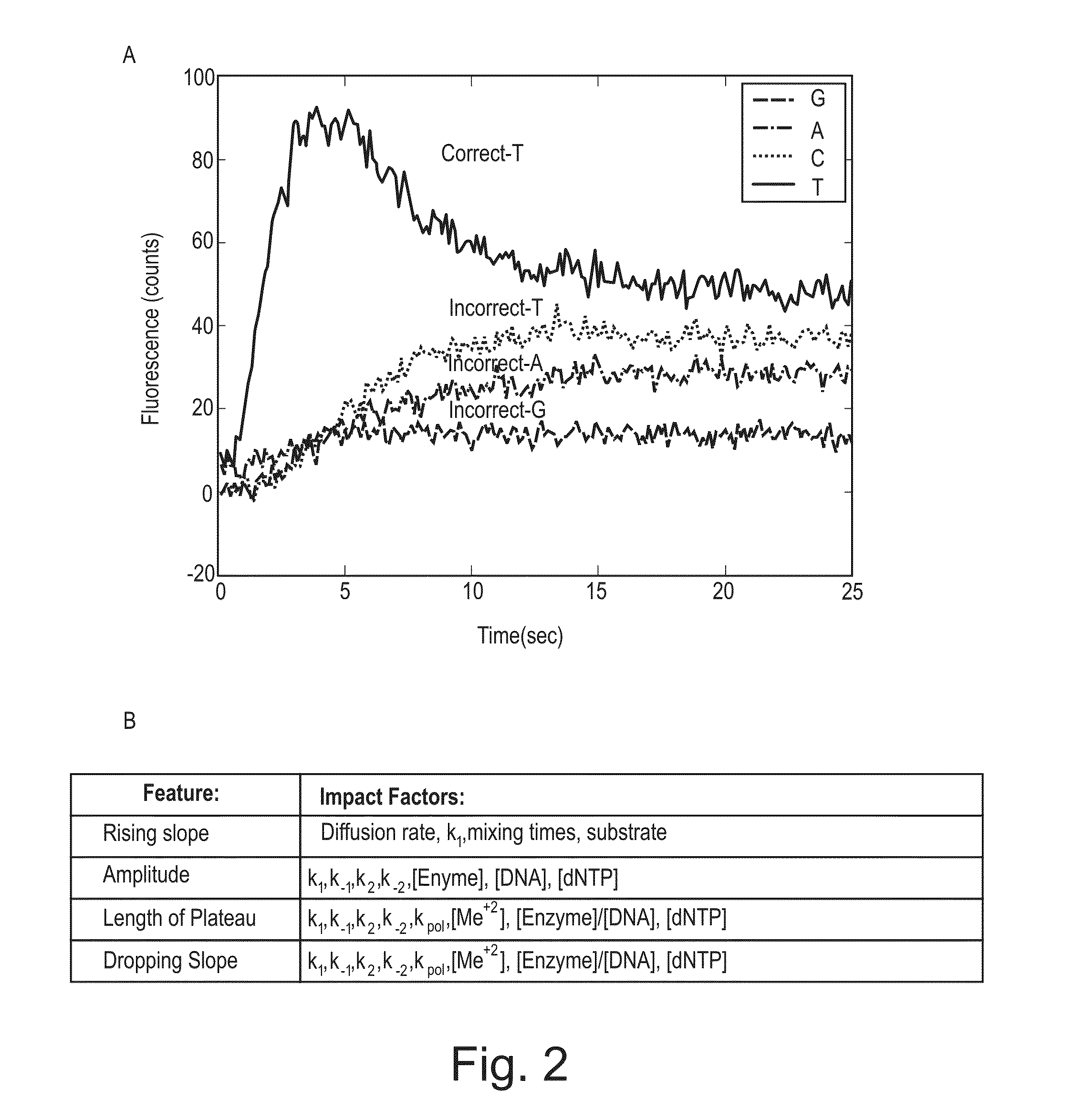
[0145]Scheme 2 was carried out as shown diagrammatically in FIG. 11. The conditions were similar to those set forth above in Example I, but with the following modifications. The target DNA template was: 5′aaaaagggaaaactecttaaaccattggaaccccgttttacccccGAGACGACGCGGTAGGCGCCAG ATATGCGATCC3′ (SEQ ID NO: 2) having the insertion sequence of GGGGGTAAAAC (SEQ ID NO: 3), and 10 μM 1-Thio-Guanosine-5′-Triphosphate (TriLink Cat #N-8007) was used in the positive reaction. Data was collected on the single molecule detection system described in Example I.
[0146]Line plots of time trace data from 225 single molecule time traces (positive and negative controls, FIG. 12) demonstrated the ability to distinguish a homopolymer pattern using the long insertion event from the modified T nucleotide bookmarked by shorter event homopolymer patterns and the insertion event of a base labeled Cy5-dCTP nucleotide. The terminal event is marke...
example iii
Ensemble-Level Detection of Pre-Equilibrium Kinetics Using Conditions from Single Molecule Schemes
[0147]Four flows of sequencing were carried out on an Illumina GA as follows.
[0148]Genome Analyzer flow cells were coated with primer and hybridized to a control template (Broad Template 3, from the Broad Institute, Cambridge, Mass.) and cluster amplification was performed according to the manufacturer's protocol using paired end cluster chemistry and paired end flow cells V4. Clusters were grown from 0.5 pM input DNA concentration. The resulting flow surface contained a monotemplate field of features for which the first insertion base was “T”. Polymerase reaction buffer was prepared from the following: 50 mM ACES pH 6.8; 150 mM NaCl; 5 mM DTT; 10 mM MgSO4; 2.5 mM 2-nitrobenzoic acid; and 0.02% Tween 20. Klenow polymerase was labeled with Cy3 as set forth in Example I. For sequencing, the Cy3 labeled Klenow polymerase was diluted to 150 nM in the polymerase reaction buffer and 4 indepen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com