Methods and compositions for identifying or quantifying targets in a biological sample
a biological sample and target technology, applied in the field of biological sample target identification or target quantification, can solve the problems of inability to augment protein information with cytometry, methods that are not well suited to the discovery of novel cell populations, and facs/scrna-seq approaches suffer from relatively low throughpu
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Design and Validation of Antibody-oligo Complexes
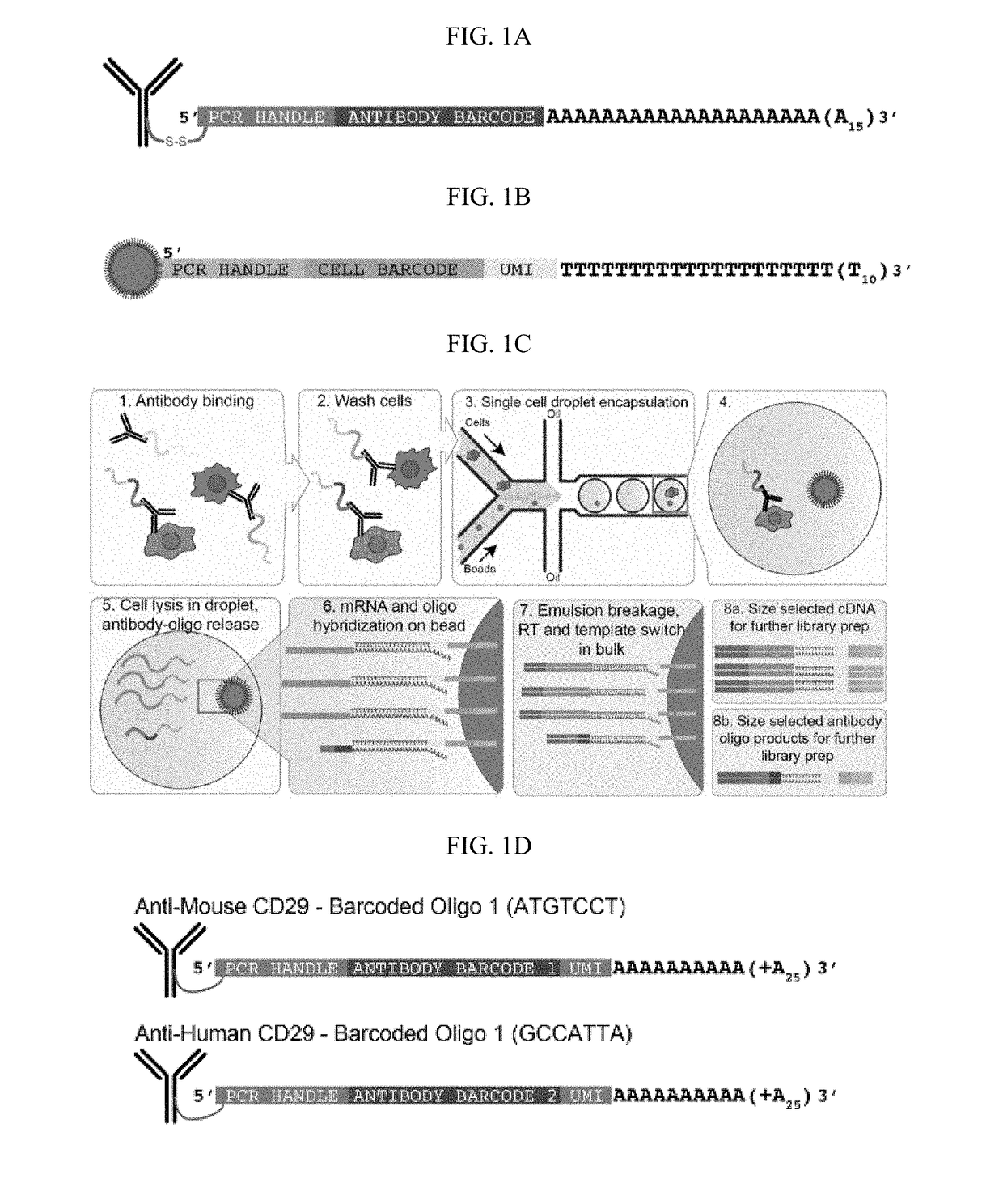
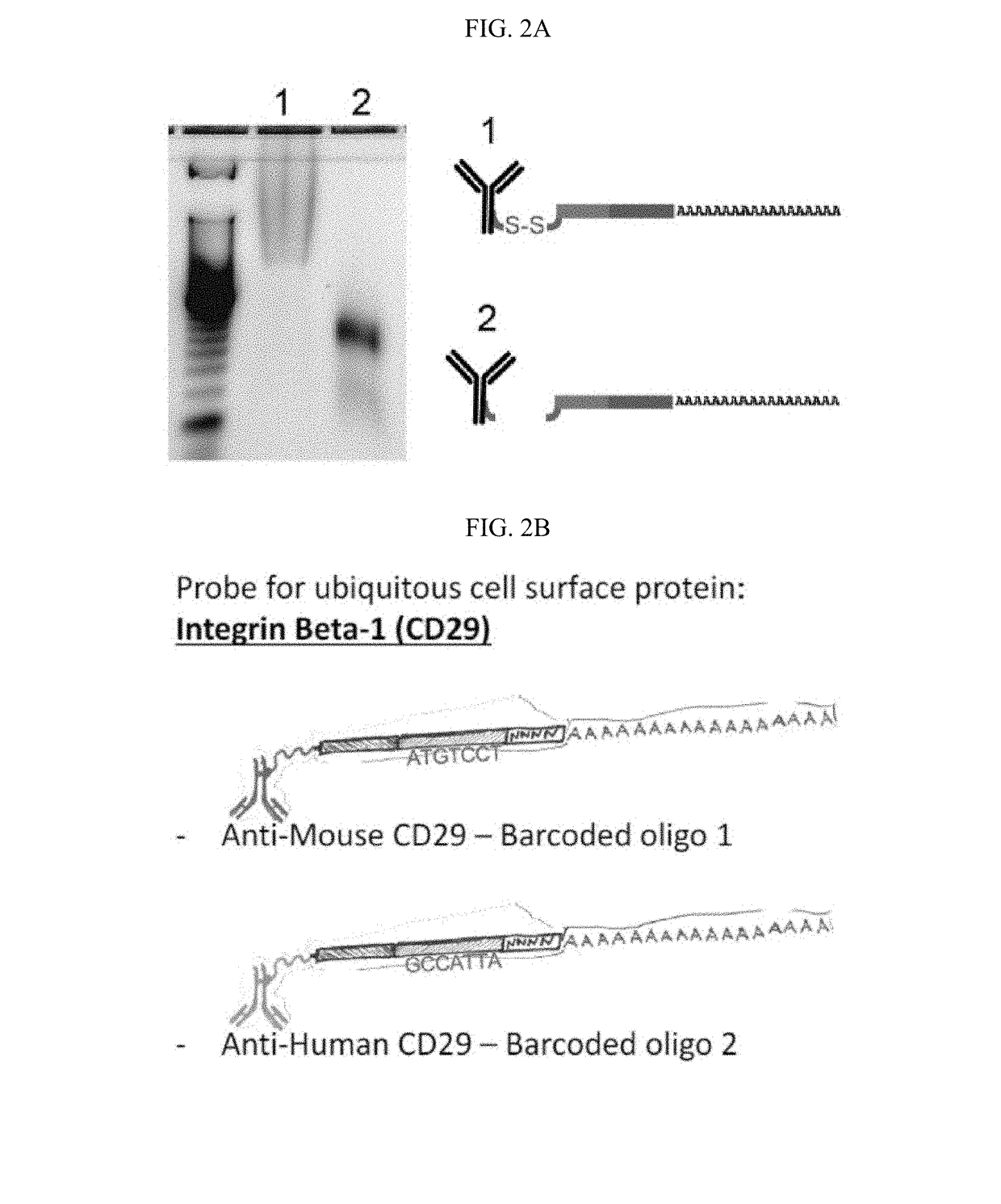
[0391]Antibody-oligos were designed with the following characteristics: a generic Amplification Handle (PCR handle) for next-generation sequencing library preparation, a unique Barcode sequence specific for each antibody, and a polyA stretch at the 3′ end (FIG. 1A). Two antibody-oligos were generated. Anti-Mouse Integrin Beta-1 (CD29) antibodies were linked to Barcoded oligo 1 containing a disulfide bridge, a common sequence (Amplification Handle, PCR handle), a unique antibody identifier Barcode (5′-ATGTCCT-3′) and a UMI containing 4 nt followed by a polyA tail (FIG. 2B, top panel). Anti-human CD29 antibodies were linked to Barcoded oligo 2 containing a disulfide bridge, a common sequence (Amplification Handle, PCR handle), a unique antibody identifier Barcode (5′-GCCATTA-3′) and a UMI containing 4 nt followed by a polyA tail (FIG. 2B, bottom panel).
[0392]For the experiments presented in Examples 1 to 7, the oligos were modified with...
example 2
Methods and Materials
[0393]Conjugation of Antibodies to DNA-barcoding Oligonucleotides. Highly specific, flow-cytometry-tested monoclonal antibodies were conjugated to oligonucleotides containing unique antibody-identifier sequences and a polyA tail. We adopted a commonly used streptavidin-biotin interaction to link oligos to antibodies19. Antibodies were streptavidin labeled using the LYNX Rapid Streptavidin Antibody Conjugation Kit (Bio-Rad, USA) according to manufacturer's instructions with modifications. Specifically, we labeled 15 μg of antibody with 10 μg of streptavidin. At this ratio, an average of two streptavidin tetramers will be conjugated per antibody molecule, which results in an average of eight binding sites for biotin on each antibody. DNA oligonucleotides with a 5′ amine modification were purchased at IDT (USA) and biotinylated using NHS-chemistry according to manufacturer's instructions (EZ Biotin S-S NHS, Thermo Fisher Scientific, USA). The optional disulfide bon...
example 3
Identification of Different Species in Mixing Sample
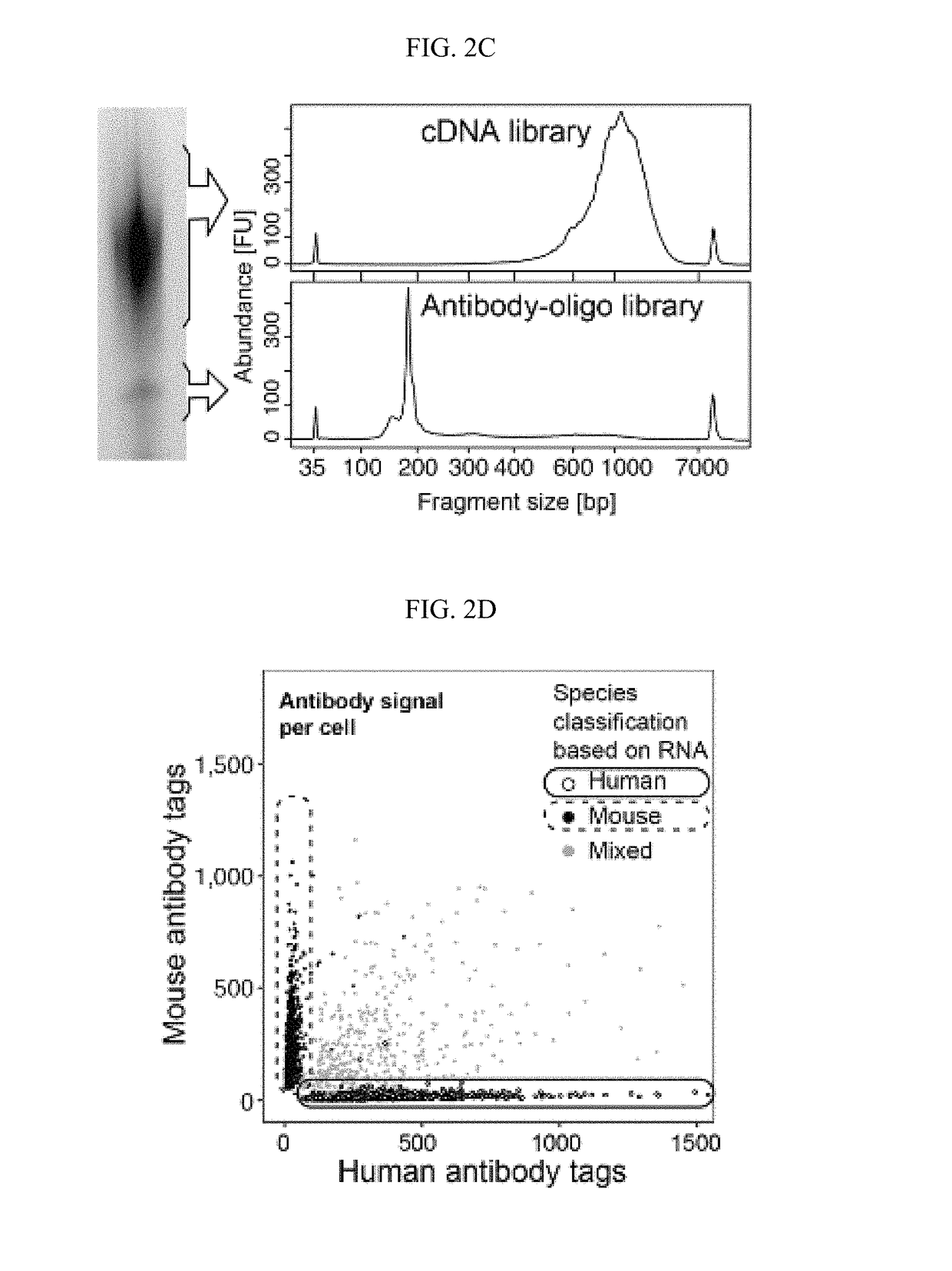
[0429]The antibody-oligo complexes described in Example 1 were incubated with cells using conditions established for flow cytometry, such as ref 22. The cells were washed to remove unbound antibodies, then single cells were encapsulated into nanoliter-sized aqueous droplets in a microfluidic apparatus designed to perform Drop-seq1 (FIG. 1C). After cell lysis (which happened immediately in the droplets when the lysis buffer contacted cells), cellular mRNAs annealed to polyT containing Drop-seq beads (FIG. 1B) via their polyA tail (FIG. 1C#6). Oligos from the antibodies also annealed to the Drop-seq beads via their poly-A stretch at the 3′ end. A unique Barcode sequence on the Drop-seq bead indexed the transcriptome of each co-encapsulated cell. After breaking the emulsion and removing the oil, reverse transcription extended the Barcoded oligo to create the first-strand cDNA from both mRNA and antibody-derived oligo templates. The cD...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Mass | aaaaa | aaaaa |
| Length | aaaaa | aaaaa |
| Length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com