Significance analysis of microarrays
a microarray and significance analysis technology, applied in the field of statistical analysis of gene related data, can solve the problems of erroneously identifying genes with changes of statistical significance, deemed potentially significant genes with scores greater than an adjustable threshold,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
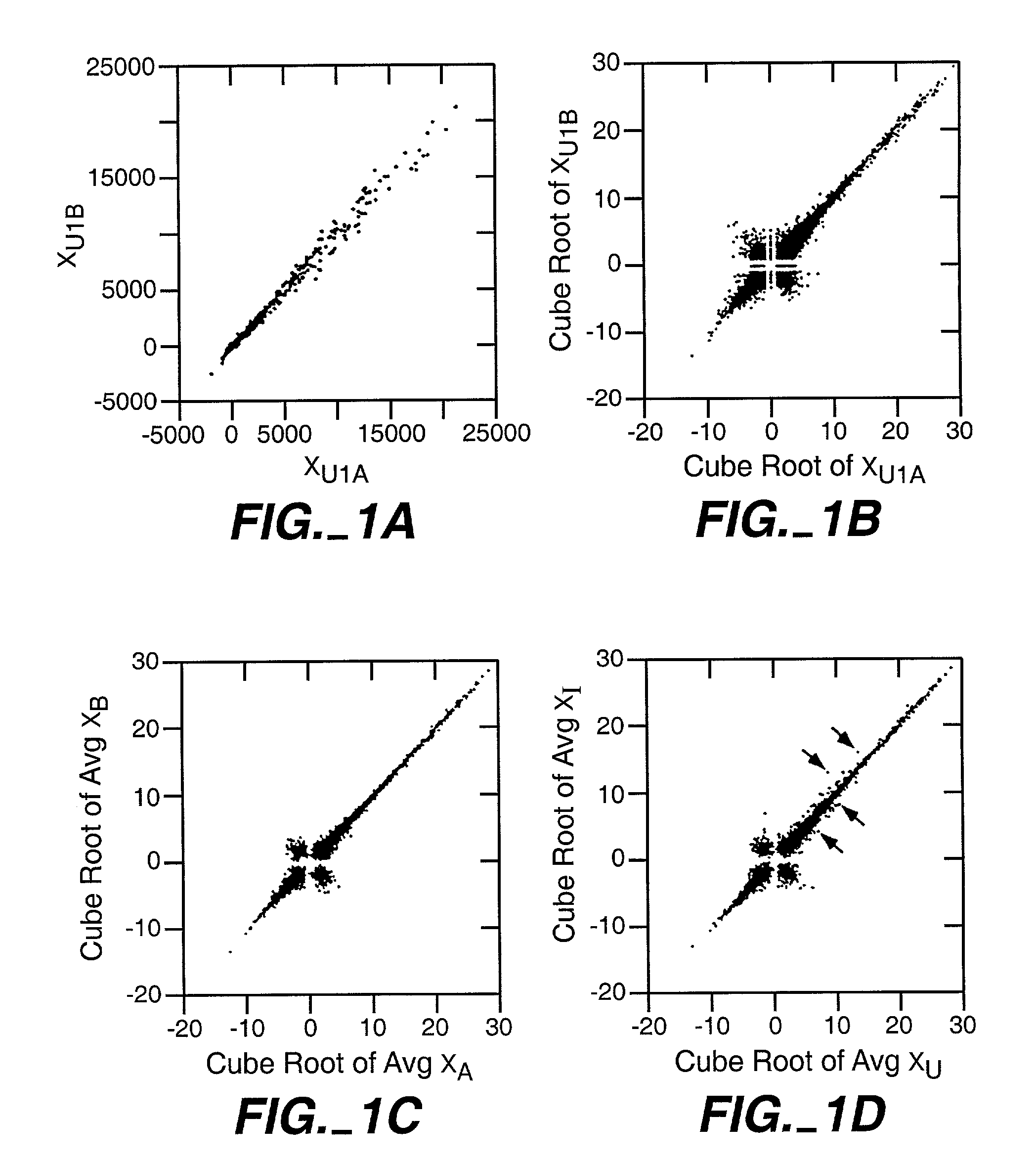
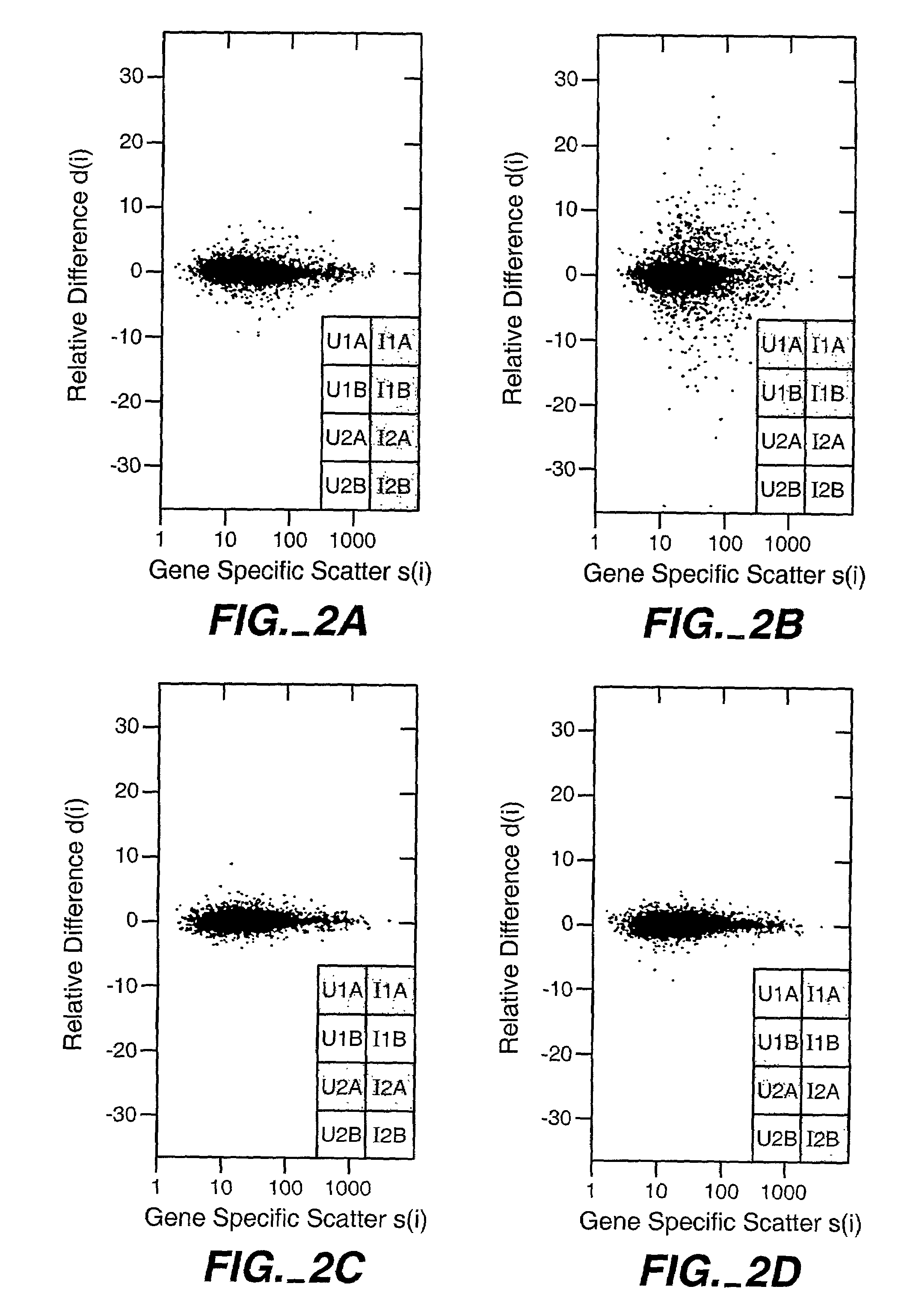
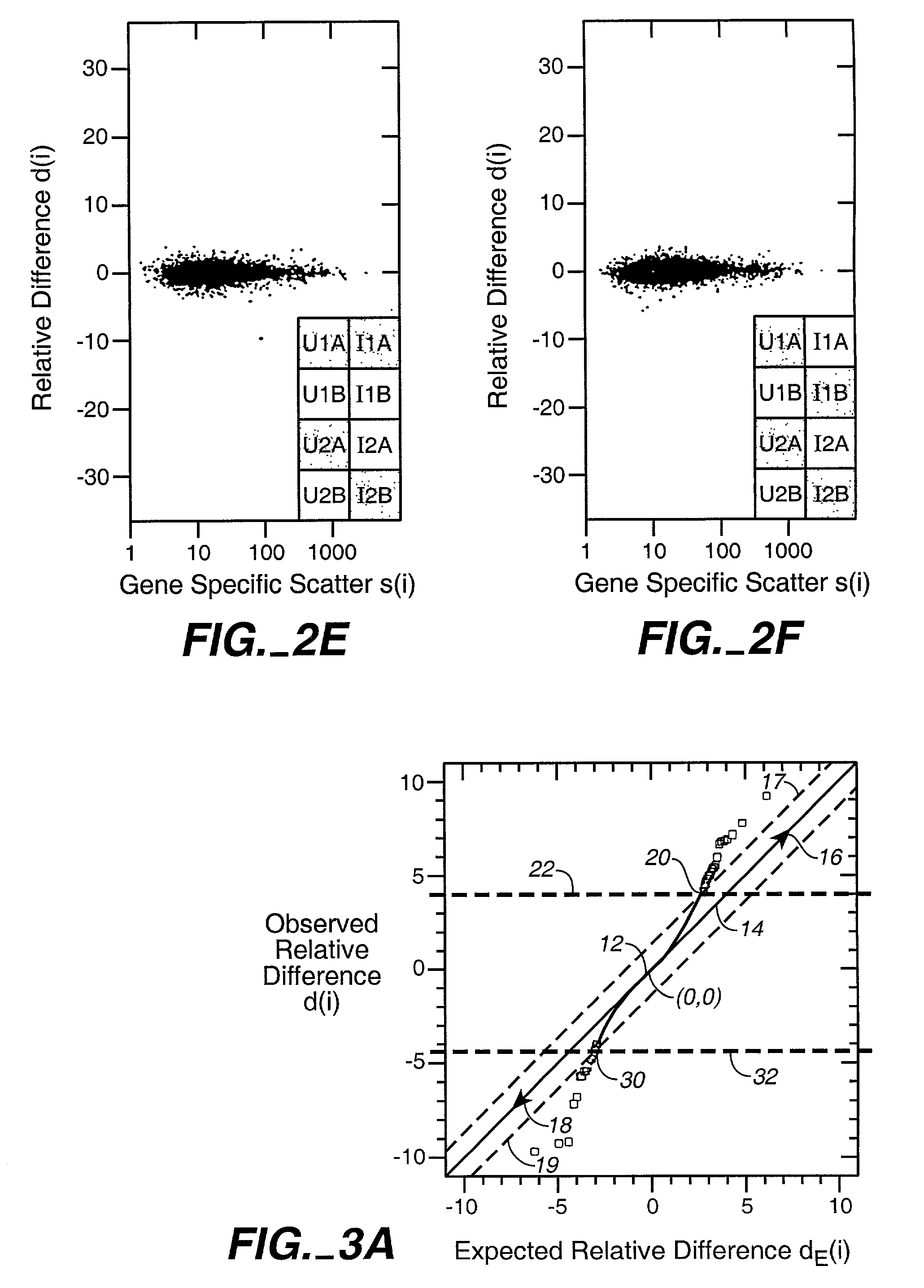
[0048]Because of its biological importance, SAM is applied to the transcriptional response of lymphoblastoid cells to ionizing radiation (IR). Although the data were obtained from oligonucleotide microarrays representing 6800 genes, SAM can also be applied to cDNA microarrays in a similar manner.
Materials and Methods Used in the Invention
[0049]Preparation of RNA. Lymphoblastoid cell lines GM14660 and GM08925 (Coriell Cell Repositories, Camden, N.J.) were seeded at 2.5×105 cells / ml and exposed to 5 Gy 24 hours later. RNA was isolated, labeled and hybridized to the HuGeneFL GeneChip® microarray according to manufacturer's protocols (Affymetrix, Santa Clara, Calif.).
[0050]Microarray hybridization. Each gene in the microarray was represented by 20 oligonucleotide pairs, each pair consisting of an oligonucleotide perfectly matched to the cDNA sequence and a second oligonucleotide containing a single base mismatch. Because gene expression was computed from differences in hybridization to ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| time | aaaaa | aaaaa |
| density | aaaaa | aaaaa |
| fluorescence | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com