Recommbined D-amino acid oxidase
一种氨基酸、氧化酶的技术,应用在生物工程领域,能够解决比活低等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0021] Example 1 Construction of D-amino acid oxidase gene recombinant plasmid pRSET-A-DAO
[0022] According to the known 5' and 3' end sequences of the Trichomona trichomona D-amino acid oxidase gene (Gonzalez, F.J., Montes, J., Martin, F., Lopez, M.C., Ferminan, E., Catalan, J., Galan, M.A. Dominguez, A. Molecular cloning of TvDAO1, a gene encoding aD-amino acid oxidase from Trigonopsis variabilis and its expression in Saccharomyces cerevisiae and Kluyveromyces lactis. Yeast 13: 1399-1408; 1997) designed primers as follows:
[0023] 5'-NdeI (introduced NdeI restriction site):
[0024] 5’-TAGGGCTGA CATATG GCTAAAATCGTTGTTATTGGTGC-3'
[0025] (Sequence 7)
[0026] 3'-BglII (BglII restriction site introduced):
[0027]5’-TAGGGCTGA AGATCT CTAAAGGTTTGGACGAGTAAGAGC-3' (SEQ ID NO: 8)
[0028] Using the plasmid pJL (Yang Yunliu et al., Chinese patent application publication number: CN 1371999A) as a template, using the above two primers, under the action of Pfu DNA polymerase...
example 2
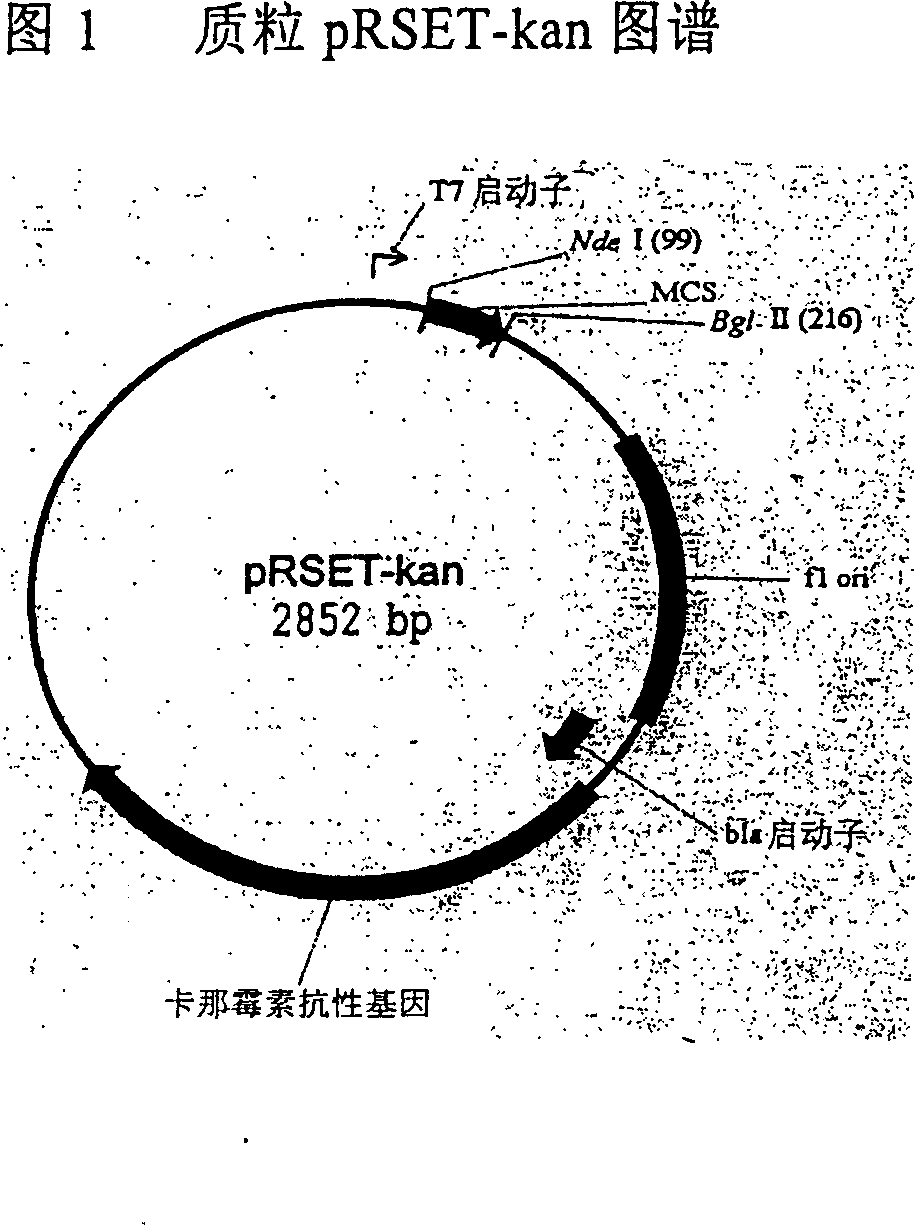
[0034] Construction of example 2 pRSET-kan vector
[0035] In order to remove the ampicillin resistance gene from pRSET-A, primers were designed according to the sequence of the vector pRSET-A as follows:
[0036] VET-F: 5'-CTGTCAGACCAAGTTTACTCATATATACTTTAG-3'
[0037] (Sequence 9)
[0038] VET-R: 5'-ACTCTTCCTTTTTCAATATTATTGAAGC-3' (SEQ ID NO: 10)
[0039] For amplifying the kanamycin resistance gene from the carrier pET-28b (Novogen), the primers are designed as follows according to the sequence of the carrier pET-28b (Novogen):
[0040] KAN-F: 5'-ATGAGTCATATTCAACGGGAAAC-3' (SEQ ID NO: 11)
[0041] KAN-R: 5'-TTAGAAAAACTCATCGAGCATCAAATG-3' (SEQ ID NO: 12)
[0042] The PCR conditions for amplifying the pRSET-A fragment that removes the ampicillin resistance gene are: 50ng pRSET-A, 0.4μM VET-F, 0.4μM VET-R, 50μM dATP, 50μM dTTP, 50μM dCTP, 50μM dGTP, 20mM Tris-HCl (pH 8.8), 10mM KCl, 10mM (NH 4 ) 2 SO 4 , 2mM MgSO 4 , 0.1% Triton X-100, 2.5U Pfu DNA polymerase, adjust t...
example 3
[0047] Example 3 Construction of recombinant D-amino acid oxidase GHA
[0048] The recombinant D-amino acid oxidase GHA was constructed by site-directed mutagenesis. The site-directed mutagenesis technique mainly refers to the description in the book PCR Protocols (Editors: John M.S. Bartlett and David Stirling. Totowa, N.J.: Humana Press, 2003.).
[0049] According to the sequence (sequence 1) of the Trichomona trichomona D-amino acid oxidase gene cloned in Example 1, the primers are designed as follows:
[0050] Primer A: 5'TAGGGCTGA CATATG GCTAAAATCGTTGTTATTG-3' (SEQ ID NO: 14)
[0051] Primer B: 5'TAGGGCTGA AGATCT CTAAAGGTTTGGACGAG-3' (SEQ ID NO: 15)
[0052] Primer C 1: 5'-GCAGGTGCCAACTGGCTC CCG TTTTACGATGGAGGCAAG-3' (SEQ ID NO: 16)
[0053] Primer D: 5'-GAGCCAGTTGGCACCTGCCCAAGG-3' (SEQ ID NO: 17)
[0054] Primers A and B are a pair of outer primers. Primer A contains an NdeI restriction site, and some bases overlap with the 5' end sequence of the D-amino acid o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com