Methods, systems, and arrays based on correlating p53 status with gene expression profiles, for classification, prognosis, and diagnosis of cancers
A technology of gene expression profiling and p53 gene, applied in the system field of predicting disease susceptibility in patients, can solve the problems of inability to provide absolute indication of p53 function, misregulation, loss of function, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0073] The molecular configuration of p53 variants is distinct from p53 wild-type.
[0074] To further visualize molecular differences between p53 variant (mt) and p53 wild-type (wt) breast cancer tumors, high-density oligonucleotide microarrays were used to analyze a whole-body series of 257 biopsies, all of which had been previously Sequencing of variants within the p53 coding region was performed. (Bergh, J., Norberg, T., Sjogren, S., Lindgren, A. & Holmberg, L. Complete sequencing of the p53 gene provides prognostic information in breast cancer patients, particularly in relation to adjuvant systemic therapy and radiotherapy. NatMed 1, 1029-34 (1995), the contents of which are hereby incorporated into this disclosure.
[0075] The original patient treatment included fresh-frozen breast cancer tumors from a cohort based on 315 women representing 65% of all breast cancers resected in the town of Uppsala between January 1, 1987, and December 31, 1989 (Bergh et al., previousl...
Embodiment 2
[0088] A gene expression classifier to predict p53 deficiency
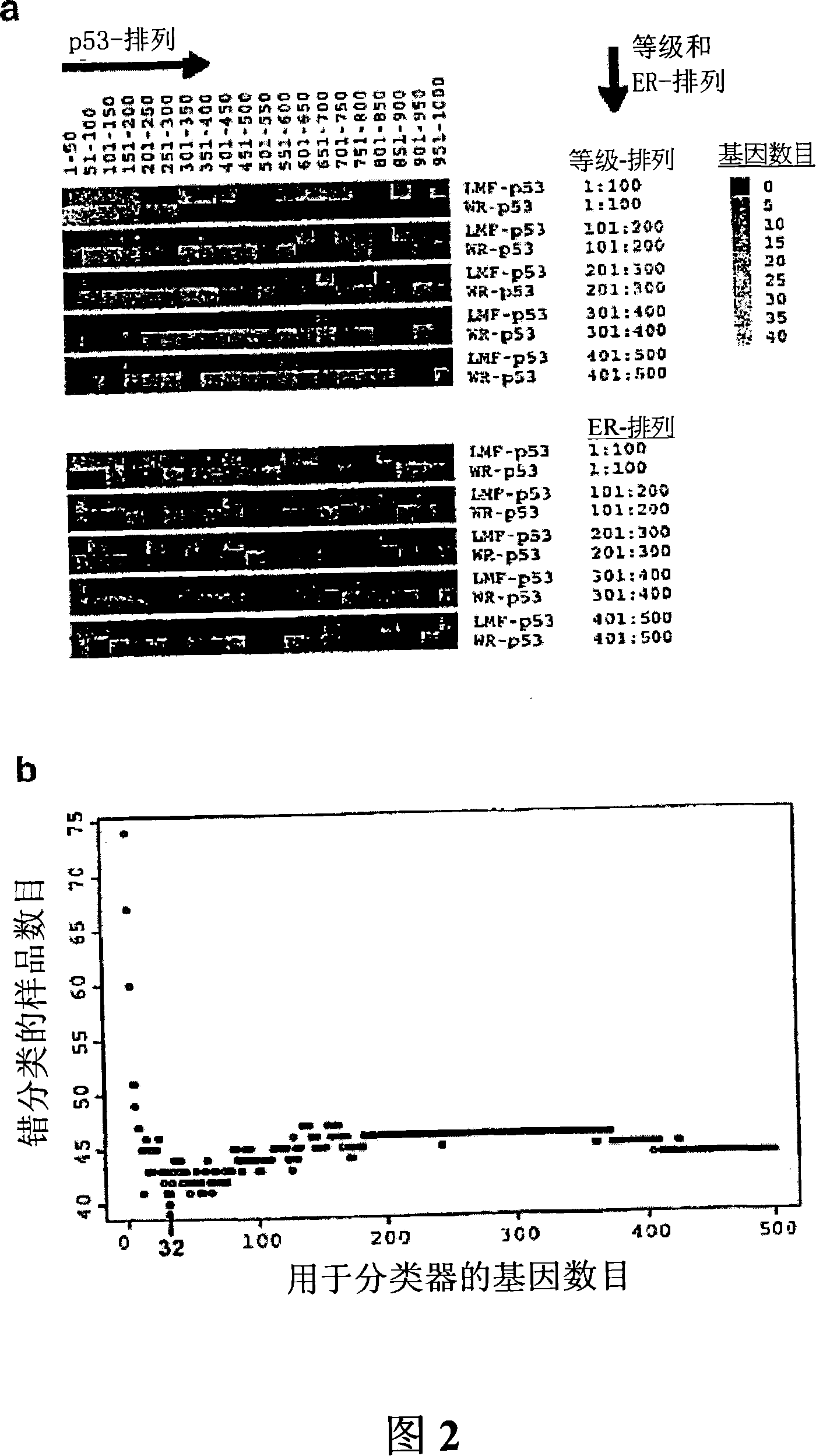
[0089] The finding that a fraction of p53wt tumors cluster with the majority of p53 mutants suggests that these tumors may in fact be p53 deficient through mechanisms other than p53 mutation. In contrast, the discovery of p53 mutants with a molecular configuration similar to most wt tumors suggests that these tumors may actually express fully functional p53. However, the assignment of tumor groups in this study was based on genes selected through a univariate permutation process and did not account for the association of p53 status with ER and grade status. This raises the possibility that, to some extent, selected genes include those that are overwhelmingly graded and / or live ER-related, thereby potentially biasing tumors toward these intrinsic properties other than p53 status Aspects produce clustering.
[0090] Therefore, a powerful gene expression-based classifier for predicting p53 status can be developed b...
Embodiment 3
[0121] The p53 classifier achieves remarkable accuracy in two independent datasets
[0122] The performance of the p53 classifier was evaluated on an independent dataset. Figure 3 shows that the classifier's genes predict p53 status in an independent cDNA microarray dataset. (A) A 9-gene subset of a 32-gene classifier predicts p53 status in an independent breast cancer dataset. The selection of the 9 genes in our classifier was based on their presence in 50% or more of the tumors. Tumors for analysis were required to have expression data for >50% of the genes. (B) An 8-gene subset of the p53 classifier predicts p53 status in an independent liver cancer dataset. Eight overlapping genes were selected based on their presence in 90% or more of the tumors. Tumors for analysis were required to have expression data for >50% of the genes. (A and B) Black vertical bars indicate p53 mutation status. Gene markers (Unigene build #167) and corresponding image clone IDs (IMAGE cloneID...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com