Primer and probe for detecting cucumber green mottle mosaic virus
A green mottled flower and leaf virus technology, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., can solve the problems of loss of edible and commercial value, discoloration of watermelon fruit pulp, economic loss, etc. To achieve the effect of high accuracy, high sensitivity and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0017] Design and synthesis of embodiment 1 primers and probes
[0018] According to the public sequences of the two virulence factors MRP (GenBank No.X64450) and EF (GenBank No.A24023) coding genes of Streptococcus suis type 2, primers and probes were designed using the probe design software Express 2.0. The primer sequences are:
[0019] CGMMV-RP (Forward): 5'-GCATAGTGCTTTCCCGTTCAC-3'
[0020] CGMMV-FP (reverse): 5'-TGCAGAATTACTGCCCATAGAAAC-3'
[0021] The sequence of the probe is: 5'-CGGTTTGCTCATTGGTTTGCGGA-3'
[0022] The 5' end of the probe was labeled with the reporter fluorescent dye FAM, and the 3' end was labeled with the quencher fluorescent dye TAMRA.
Embodiment 2
[0023] The extraction of embodiment 2 total RNA
[0024] 1) Take 0.1g of diseased leaves, cut into small pieces, grind into powder with liquid nitrogen, transfer to a sterilized 1.5ml centrifuge tube, then add 1ml of Trizol reagent, shake vigorously;
[0025] 2) Centrifuge at 12000g for 10min at 4°C to remove insoluble components, and transfer the supernatant to a new 1.5ml centrifuge tube;
[0026] 3) Keep at room temperature for 5 minutes, add 0.2ml of chloroform, shake vigorously for 15s, then keep at room temperature for 2-15 minutes, then centrifuge at 12000g for 15 minutes at 4°C;
[0027] 4) Transfer the upper aqueous phase to a new 1.5ml centrifuge tube, add 0.5ml isopropanol, invert and mix, and keep at room temperature for 15 minutes;
[0028] 5) Centrifuge at 12000g for 10min at 4°C, and the RNA will form a precipitate on the side wall and bottom of the tube;
[0029] 6) Pour off the supernatant, add 75% ethanol to wash the precipitate, then centrifuge at 7500g fo...
Embodiment 3
[0031] The establishment of embodiment 3 Real-time RT-PCR amplification method
[0032] 1. Real-time fluorescent RT-PCR reaction system
[0033] Real-time fluorescent RT-PCR reaction was performed using total RNA as a template, that is, 20.25 μL DEPC-treated water, 25 μL 2×Master Mix withoutUNG, 1.25 μL 40×MultiScribe and RNase Inhibitor Mix, 1 μL primer CGMMV-FP ( 20 μmol / L), 1 μL primer CGMMV-RP (20 μmol / L), 0.5 μL probe CGMMV-FAM (20 μmol / L), 1.6ng total RNA.
[0034] 2. Real-time fluorescent RT-PCR reaction conditions
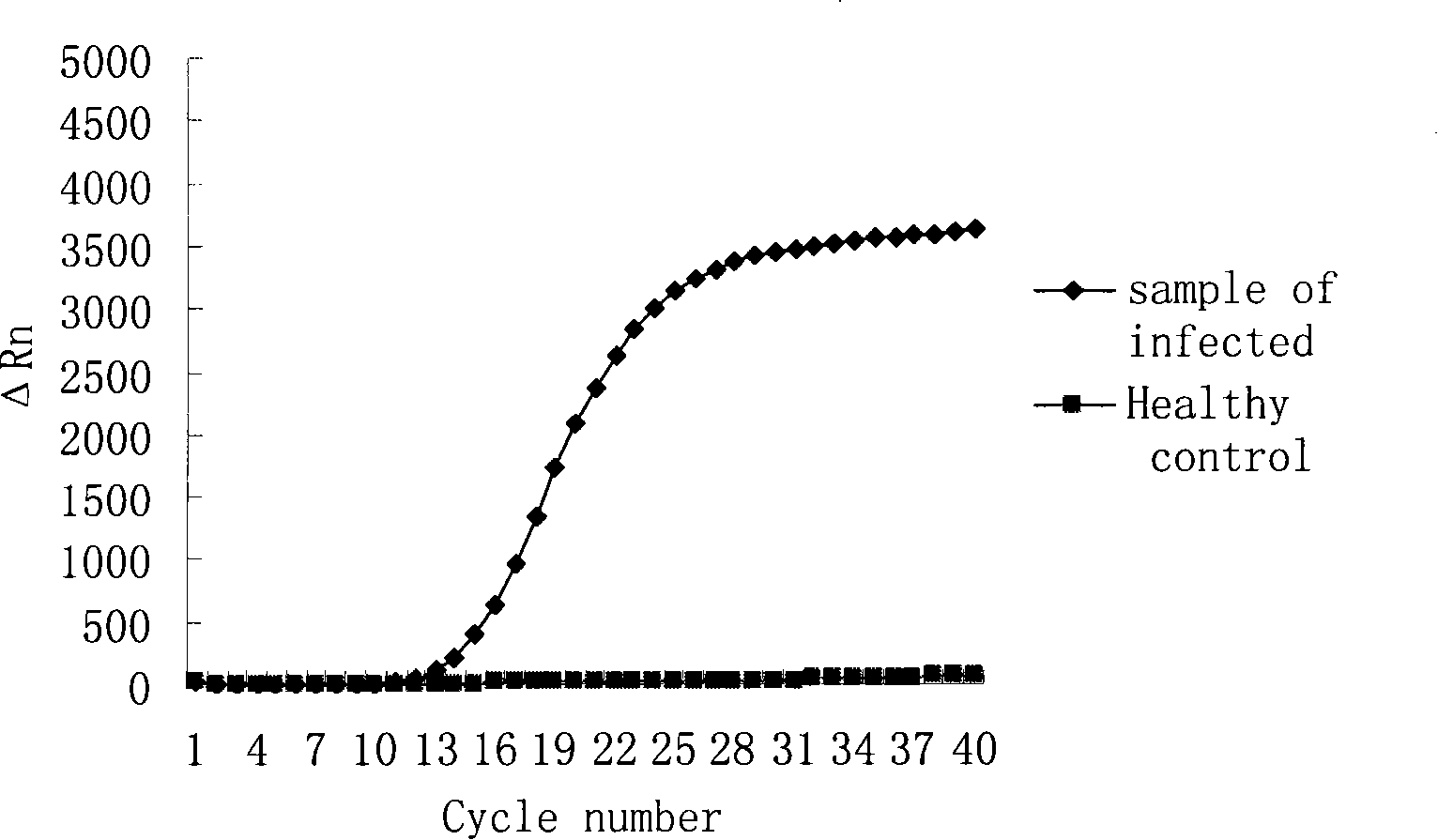
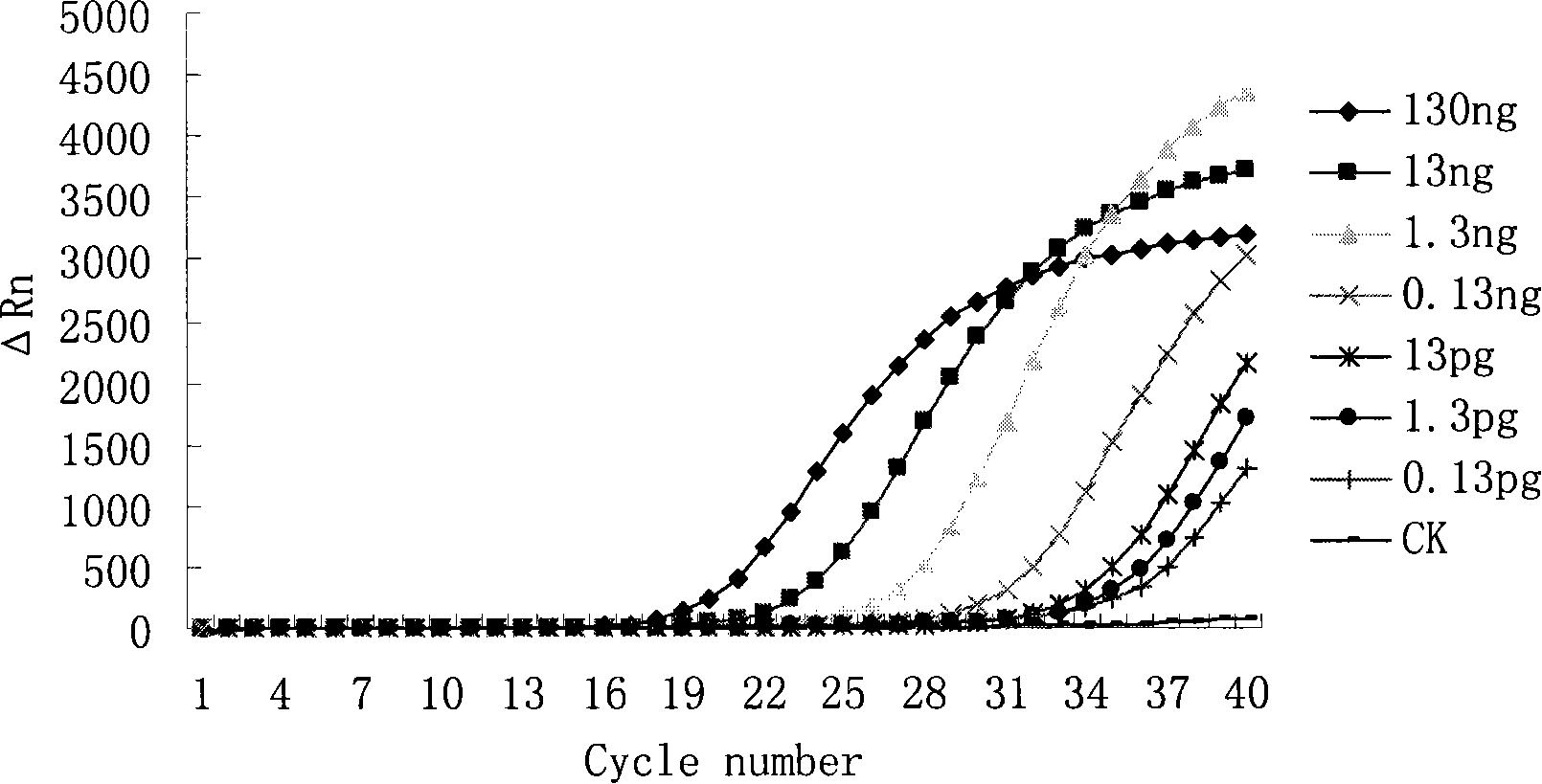
[0035] After putting the sample tube into the ABI 7700 fluorescent PCR instrument of ABI Company, set the following conditions for reaction: 42°C, 30min; 95°C, 5min; 95°C, 15s; 60°C, 1min, a total of 40 cycles. Collect data after each cycle. After the reaction, judge the result according to the amplification curve.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com