Method for rapidly extracting plants sample DNA
An extraction method and plant technology, applied in the field of molecular biology, can solve the problems of many extraction steps, troublesome, complicated, etc., and achieve the effect of simple process, many operation steps and fast process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
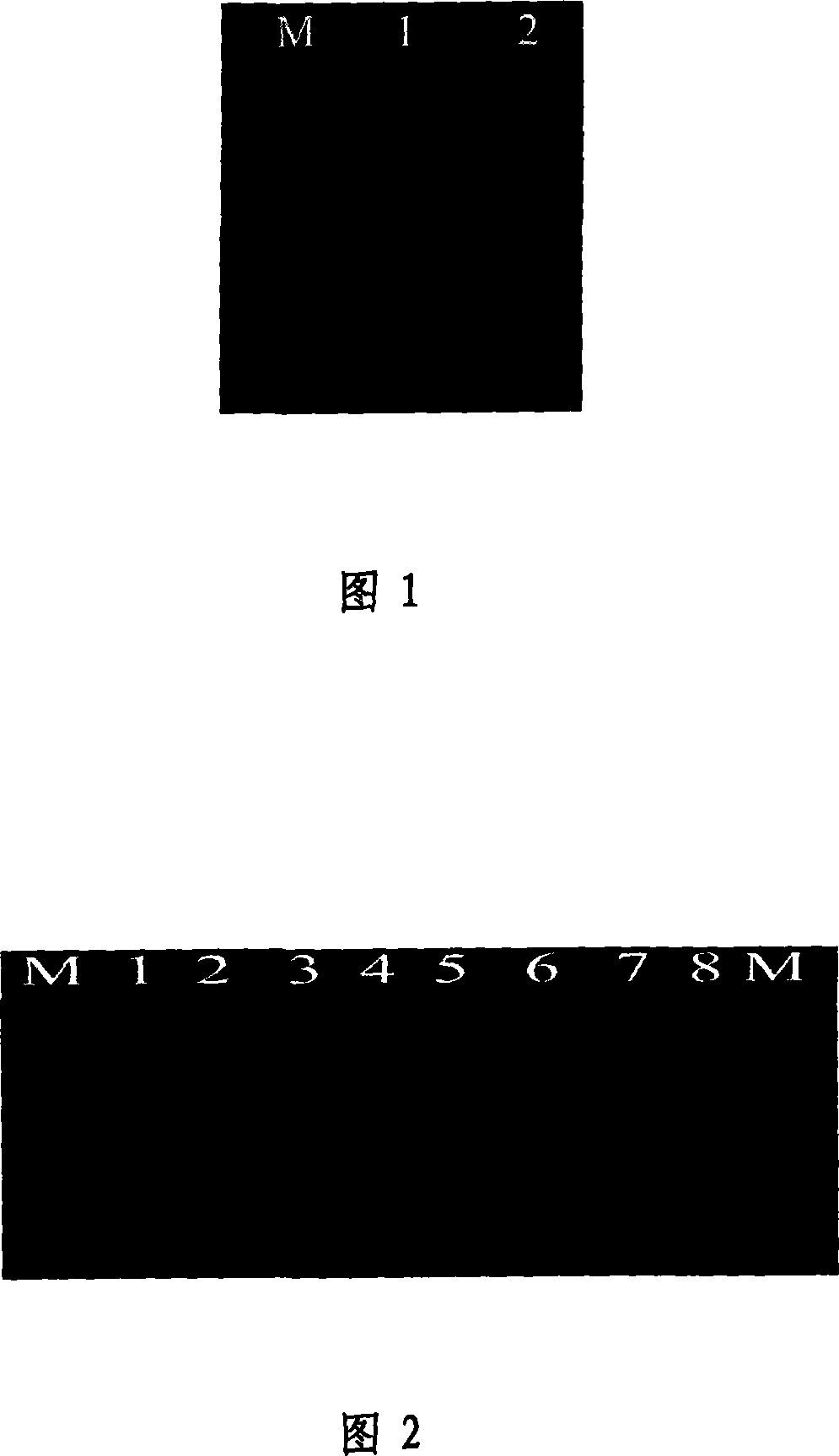
[0072] Using the rapeseed sold in the market as a sample, the method of the invention is used to rapidly extract DNA. The preparation method is as follows:
[0073] (1) Take about 100 mg of rapeseed, add a small amount of liquid nitrogen to a mortar, and grind it quickly. Add liquid nitrogen repeatedly 3 to 4 times to grind to powder, and put it into a sterilized centrifuge tube.
[0074] (2) Add 800 μL of CTAB DNA extraction buffer, vortex mix, place in a water bath at 65°C for 0.5 h, and centrifuge at 12,000 r / min for 5 min.
[0075] (3) Take the supernatant in a sterilized centrifuge tube, add 3mol / L sodium acetate solution 1 / 10 times the volume of the supernatant and 0.8 times the volume of isopropanol, centrifuge at 12000r / min for 5min, discard the supernatant .
[0076] (4) 100 μL of 10% Chelex-100 was added to the precipitate, and the precipitate was fully dissolved in the solution, centrifuged at 12000 r / min for 10 min, and the supernatant was taken and stored for P...
Embodiment 2
[0091] Taking corn grains from the farm of our hospital as samples, the method of the present invention was used to rapidly extract DNA.
[0092] (1) Take about 100 mg of corn in a mortar, add liquid nitrogen to grind to powder, and put it into a sterilized centrifuge tube;
[0093] (2) Add 800 μL cetyltrimethylammonium bromide extraction buffer, vortex mix, place in a water bath at 65°C for 40 minutes at a constant temperature, and centrifuge at 15,000 r / min for 8 minutes;
[0094] (3) Take the supernatant in a centrifuge tube, add 3mol / L sodium acetate solution 1 / 10 times the volume of the supernatant and 0.8 times the volume of isopropanol, mix well, centrifuge at 15000r / min for 8min, discard supernatant;
[0095] (4) Add 200 μL of 10% Chelex-100 solution to the precipitate, and fully dissolve the precipitate in the solution, centrifuge at 12,000 r / min for 10 min, take the supernatant, test the DNA, save it for PCR amplification.
[0096] PCR test:
[0097] 1. Primer seq...
Embodiment 3
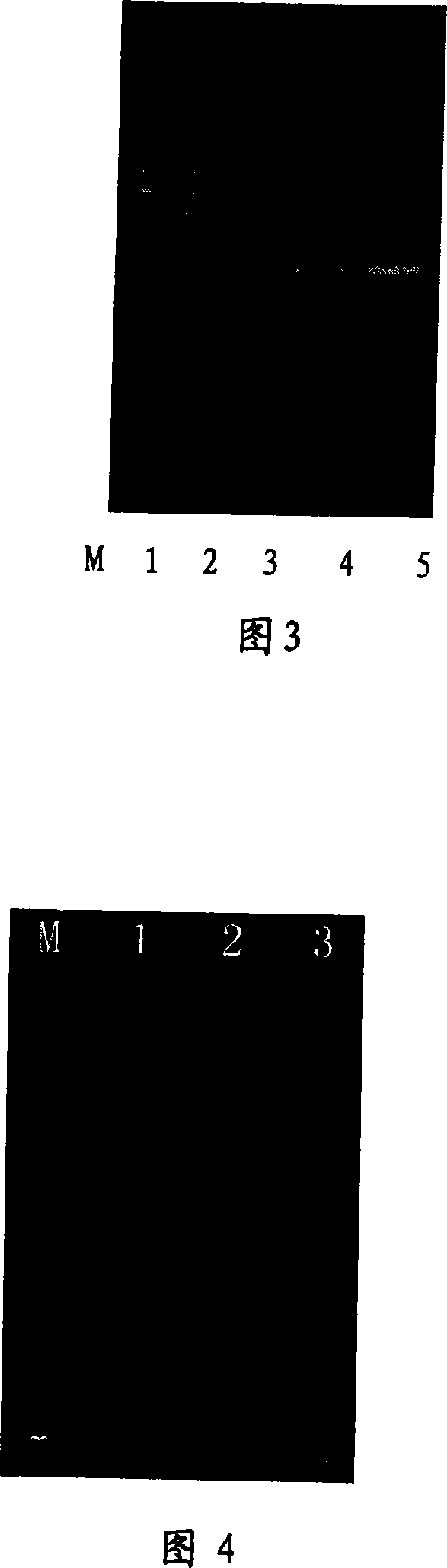
[0110] Using the soybean callus collected in our laboratory as a sample, the method of the present invention was used to rapidly extract DNA.
[0111] (1) Take about 100 mg of the sample, add liquid nitrogen to the mortar, grind it to powder, and put it into a sterilized centrifuge tube;
[0112] (2) Add 600 μL cetyltrimethylammonium bromide extraction buffer, vortex mix, place in a water bath at 65°C for 40 minutes, centrifuge at 12,000 r / min for 10 minutes;
[0113] (3) Put the supernatant in a centrifuge tube, add 1 / 10 times the volume of 3mol / L sodium acetate solution and 0.8 times the volume of isopropanol, mix well, centrifuge at 12000r / min for 10min, discard the supernatant;
[0114] (4) Add 100 μL of 10% Chelex-100 solution to the precipitate, and fully dissolve the precipitate in the solution, centrifuge at 15000 r / min for 10 min, take the supernatant, save it, and use it for PCR amplification.
[0115] 1. Composition of PCR reaction system:
[0116] 10× PCR reactio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com